Isolating RNA from hemocytes
Oct. 30, 2020
Samples:
| Animal_ID | Condition | Sex | Stage | Location |
|---|---|---|---|---|
| 3 | ambient | F | mature/spent | Rack 6, col 2, row 2 |
| 6 | ambient | F | ripe | Rack 6, col 2, row 2 |
| 7 | ambient | F | late active | Rack 6, col 2, row 2 |
| 015 | low pH | F | early active | Rack 6, col 2, row 2 |
| 025 | low pH | F | late active | Rack 6, col 2, row 2 |
| 053 | ambient | F | early active | Rack 6, col 2, row 2 |
| 057 | ambient | F | ripe | Rack 6, col 2, row 2 |
| 060 | ambient | F | early active | Rack 6, col 2, row 2 |
RNeasy Protocol Modificaitons:
- Sample notes:
- These samples were supposed to be pelleted hemocytes from hemolymph samples, however pellets were not really visible in most samples
- this is not totally surprising because they are usually transparent/translucent cells anyways.
- Some samples had some other tissue type in them
- 015 has a green pellet
- 025 had a yellow pellet
- These samples were supposed to be pelleted hemocytes from hemolymph samples, however pellets were not really visible in most samples
- generally following what I did yesterday https://shellywanamaker.github.io/380th-post/
- add BME to lysis buffer
- Made 7mL RLT + 70uL BME (-20C 2nd shelf drawer 7)
- Added 350uL RLT + BME to pellets, vortexed and pipetted up and down to mix
- Pipetted lysate into Qiashredder colums and spun at max speed for 2 min.
- Transfered lysate to gDNA eliminator column and spun for 30 seconds at 11000 rpm
- Saved 180uL of homogenized lysate and froze at -80C in hemolymph box (Rack 6, col. 2, row 2)
- Followed protocol for purification of total RNA
- Added 270uL of 100% ethanol to ~180uL homogenized lysate and transfered to minElute column
- spun at 11000 rpm for 15 sec.
- Added 500uL buffer RPE and spun at 11000rpm for 15 sec. Repeated this step for a total of 3 times
- Added 500 uL buffer RPE and spun at 11000 rpm for 2 minutes
- dried the RNeasy minElute column by spinning at max speed for 5 minutes with the lids open and using a new collection tube
- eluted RNA in 15uL RNase free H2O by spinning at 11000 rpm for 1 minute
- I saved the columns
Quantification
- I measured the concentrations of the RNA on the nanodrop because the Qubit was being used.
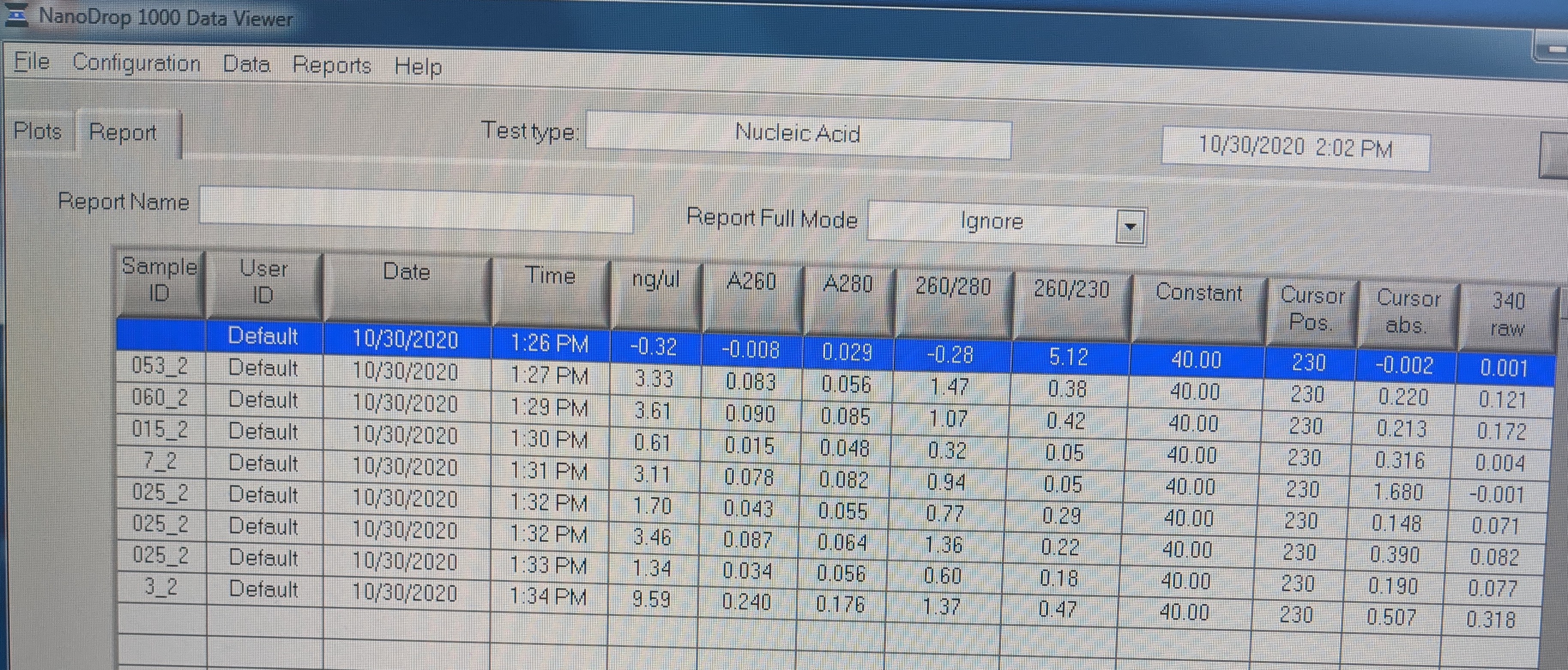

- *NOTE: Not all Sample IDs in image are correct and samples are listed as follows:
- H2O
- 053_2
- 060_2
- 015_2
- 7_2
- 025_2
- 057_2
- 6_2
- 3_2
- eluted off the RNeasy minElute columns again with 15uL of warmed RNase free H2O (~37C) into tubes labeled with just the sample numbers in Sharpie.
- All RNA samples were stored in the -80C box Rack 6, Col. 2, Row 2
- discussed these results in Science hour and decided to check qubit concentrations because nanodrop can be inaccurate when RNA is at low concentration
- Also came up with a plan to check samples for RNA by doing a qPCR on the RNA
Nov. 3, 2020
Quantification of Oct. 30 RNA
- thawed RNA on ice
- checked 1uL of each sample
- all samples are too low for quantification
- Standard 2 = 10.4ng/uL (should be 10ng/uL)
RNeasy Protocol Modificaitons:
I decided to go back to the homogenized lysates that I save on Oct. 30 to see if I could get any RNA from them.
- I repeated the homogenization and gDNA elimination steps to rule out too much tissue or clogged columns
- I thawed homogenized lysates at 37C for 4 minutes
- gave them a quick spin
- added 350uL RLT buffer (no BME)
- Pipetted up and down to mix and transfered to Qiashredder columns, spun at max speed for 2 minutes
- Transfered lysate to gDNA eliminator columns and spun for 30 seconds at max speed
- Followed same protocol as Oct. 29:
- Added 530uL 70% EtOH (since I had 350uL of lysis buffer + 180uL of homogenate), pipetted up and down to mix and transferred to minElute columns. Spun at 9000 x g for 15 seconds, discarded flow-through
- Added remaining homogenized lysate to column, spun at 9000 x g for 15 seconds and discarded the flow-through
- Added 700uL of buffer RW1 to minElute column and spun for 15 sec at 9000 x g, discarded flow-through
- Added 500uL buffer RPE to minElute column and spun for 15 sec at 9000 x g, discarded flow-through
- added 500uL 80% EtOH to column, spun for 2 minutes at 9000 x g, and discarded flow-through.
- Dried column by placing it in new collection tube, and spinning for 5 minutes on max speed with the column lids open
- Eluted with 20uL RNase free H2O prewarmed to 65C for 10 min on the bench. Then spun at max speed for 2 minutes.
Quantification
- checked 1uL of each sample
- all samples are too low for quantification
- Standard 2 = 9.92 ng/uL (should be 10ng/uL)
Conclusions
- It seems these samples may not have had very many cells at all since there is no RNA, because I was able to successfully isolate RNA from an old hemocyte sample from Nov 2018 but not these samples.
- My hunch is that these hemolymph samples are very variable and I’m don’t feel confident they contain sufficient hemocytes to do this experiment.
- I still have the “lymph” part of these samples, but if it’s just seawater then they’re a little useless.
- I wonder if there is a way to tell, or if these could be used for the SRM assay Emma developed?
- I also still have siphon, ctenidia, and gonad for these animals so it’s still possible to do develop qPCR on gonad tissue, but the whole idea was for this non-lethal sampling.
- since it’s possible to do gonad biopsies, this might be worth it? But will discuss with the FFAR group. We never tested how non-lethal the gonad biopsies were, particularly from our attempt with an 18G needle in January 2020
- Will consult with Steven and the group on how to proceed