Isolating RNA from 2018-19 hemolymph samples
I decided to use the Qiagen RNeasy micro prep kit because the max capacity is 45ug and you can elute in as little as 10uL. The Zymo microprep DNA/RNA kit columns have a max binding capacity of 5ug DNA and 10ug RNA. One concern I had not knowing how many cells are in these sample was overloading the column which can lead to low yield, impure RNA, and inefficient isolation.
To test the protocol out, I used a couple of old samples that are not part of the experiment. These were hemolymph samples from Nov. 27 2018 from two adults that I reared at UW.
Prep work: Made 80% ethanol (4mL RNase free H2O + 1mL 100% EtOH) and 70% ethanol (3.5mL RNase free h2O + 1.5mL 100% EtOH).
I followed the standard protocol as follows:
- Added 350uL Buffer RLT plus to pellets, vortexed
- one sample (“star”) has visible tissue in the pellet that didn’t full dissolve
- Pipetted lysate onto QIAshredder spin column and spun at max speed for 2 min.
- Transfered homogenized lysate to a gDNA eliminator spin column and spun at max speed for 30 sec.
- I reserved 175uL of the homogenized lysate after gDNA elimination in an eppy and labeled it as “Chewy 11/27 RNeasy HM” and “Star 11/27 RNeasy HM” and saved in -80C in Rack 5, Row 1, column 3. This was in case I screwed up on the prep that I’d have something to go back to.
- Added 200uL of 70% EtOH to flow through, pipetted up and down to mix, then transfered to an RNeasy MinElute spin column and spun at max speed for 15 sec.
- Added 700uL Buffer RW1 to the column and spun at max speed for 15 sec, dicarded flow through
- Added 500uL RPE buffer and spun at max speed for 15 sec, then discareded the flow through. I repeated this step by accident
- Added 500uL of 80% EtOH and spun for 2 min at max speed
- Transferred mini column to clean collection tube and spun for 3 minutes on max speed to dry column. Then remembered to open the lid and spun again for an additional 4 minutes
- Transferred column to a clean 1.7mL eppy, added 15uL RNase free H2O and spun at max speed for 1 min to elute. Pipetted the residual liquid on the edge of the column directly onto the column and spun again for 1 min.
Quantifying RNA
Qubit
- I measured 1 uL of RNA with the Qubit HS RNA kit.
- Star: too low for detection
- Chewy: 38.0 ng/uL
- Standard 2: 10.2ng/uL
Nanodrop
- Values:
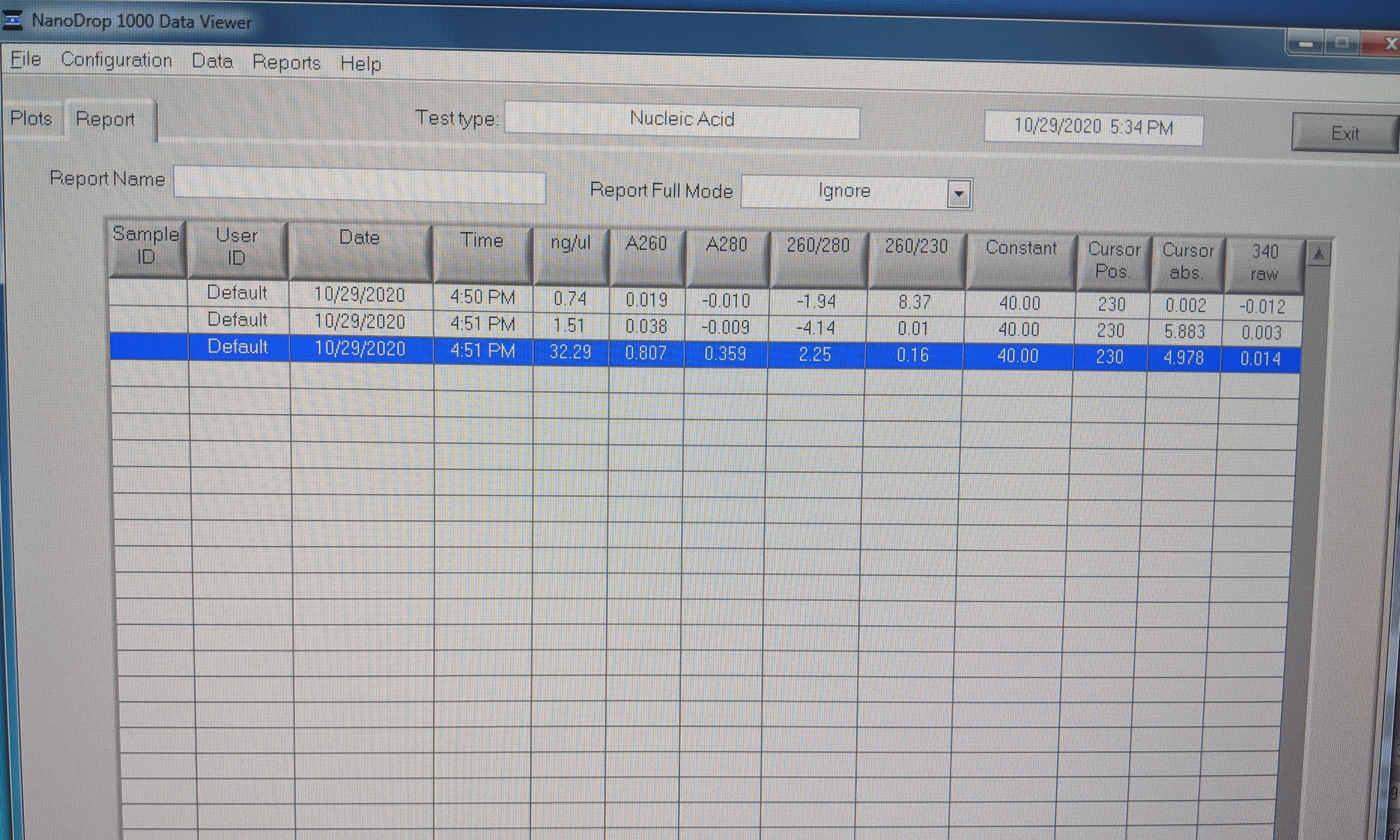
-
Plots:
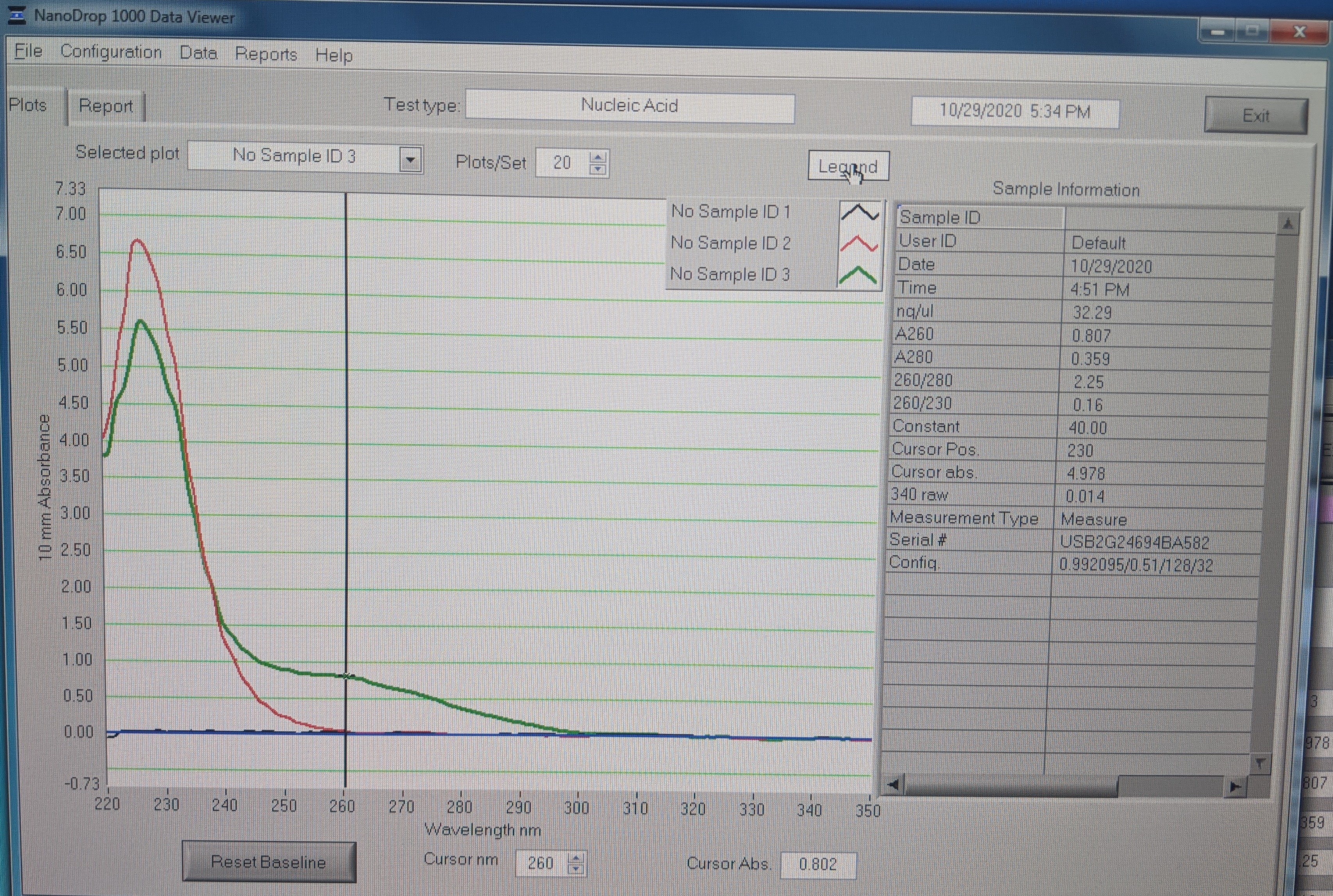
- First sample: RNase free H2O 0.74ng/uL
- Second sample: Star: 1.51ng/uL; looks like no RNA
- Third sample: Chewy: 32.29ng/uL
- 260/280 is 2.25; it should be 2.0 for pure RNA (pure DNA is 1.8). You can see the hump in the plot at 260nm.
- The 260/230 ratio is suppose to be 2 but this one is 0.16. This plus the big peak at 230nm is contamination most likely from guanidine hydrochloride salt in the RLT buffer. Qiagen says that is shouldn’t interfer with downstream reactions https://www.qiagen.com/us/resources/faq?id=c59936fb-4f1e-4191-9c16-ff083cb24574&lang=en.
- This paper https://www.microbiologyresearch.org/content/journal/jgv/10.1099/0022-1317-40-1-239?crawler=true&mimetype=application/pdf says longer centrifuge times and faster speeds promote the precipiation of guanidine HCl so I need to be more careful about how long I spin and how fast. It also mentioned EtOH washing can help reduce it, so I might try an additional 80% EtOH wash step.
Plan for tomorrow
Hemocyte samples to prep:
-
location: Rack 6, col.2, row 2:
- 053
- 060
- 015
- 7
- 025
- 057
- 3
- 6
- 8
- 14
-
location: Rack 5, col.3, row 1
- 021
- 022
- 023
- 024
- Prepare enough reagents for 20 preps:
- Add 350uL RLT/sample = 7mL RLT + 70uL B-ME (-20C 2nd shelf drawer 7)
- this will help with dissociated proteinaceous tissue and help denature RNases
- Add 350uL RLT/sample = 7mL RLT + 70uL B-ME (-20C 2nd shelf drawer 7)
- Label all tubes:
- qiashredders (14)
- gDNA eliminator columns (14)
- minElute columns (14)
- eppys (2 x 14)
- qubit (16)