Run DMRfind on new alignments
This is the follow up analysis from these posts:
- https://shellywanamaker.github.io/232th-post/
- Copy 5x.tab files to Ostrich, Convert 5x.tab files to allc format, and Run DMRfind:
- script here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200327/20200327_DMRfind.sh
- screen output here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200327/screenlog.0
- all files, scripts, and logs were copied to Gannet here:https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200327/
- 5x.tab files: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200327/merged_tab/
- allc files: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200327/
- DMRfind output:
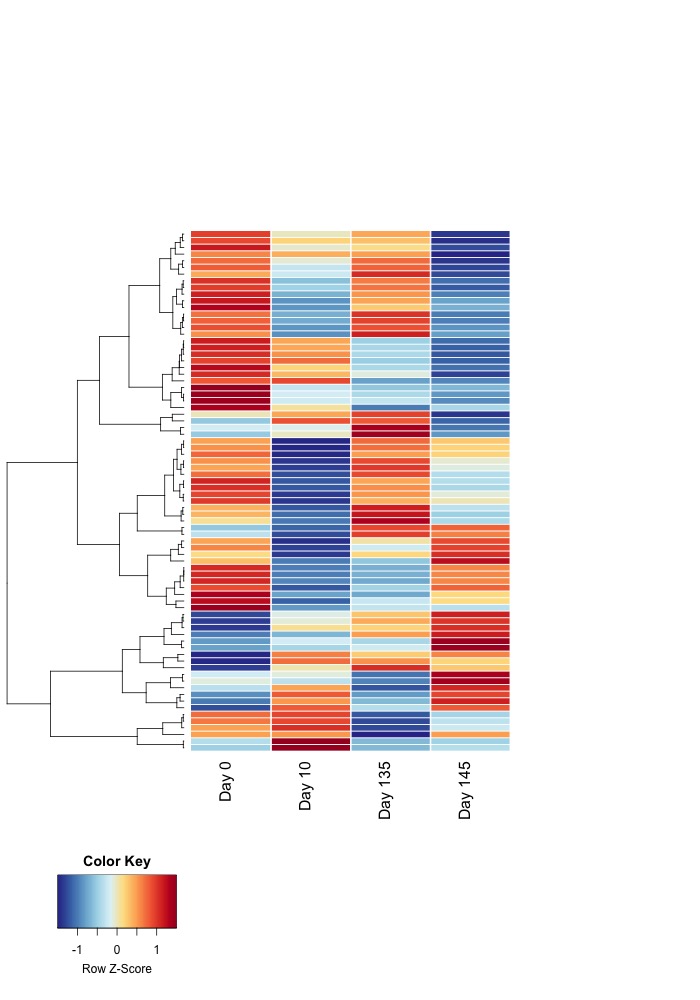
- All ambient samples:
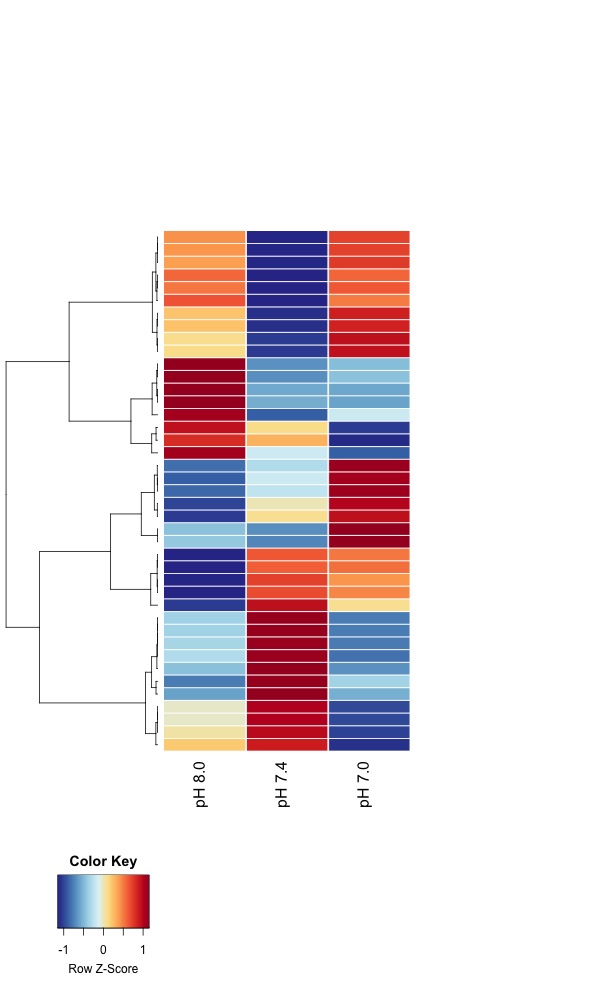
- Day 10 all pH samples:
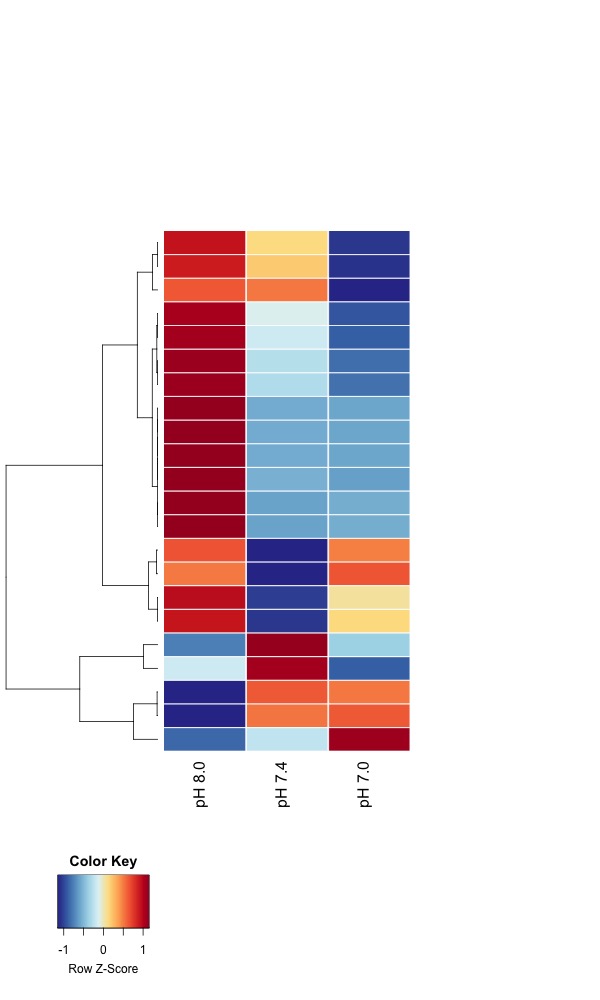
- Day 135 all pH samples:
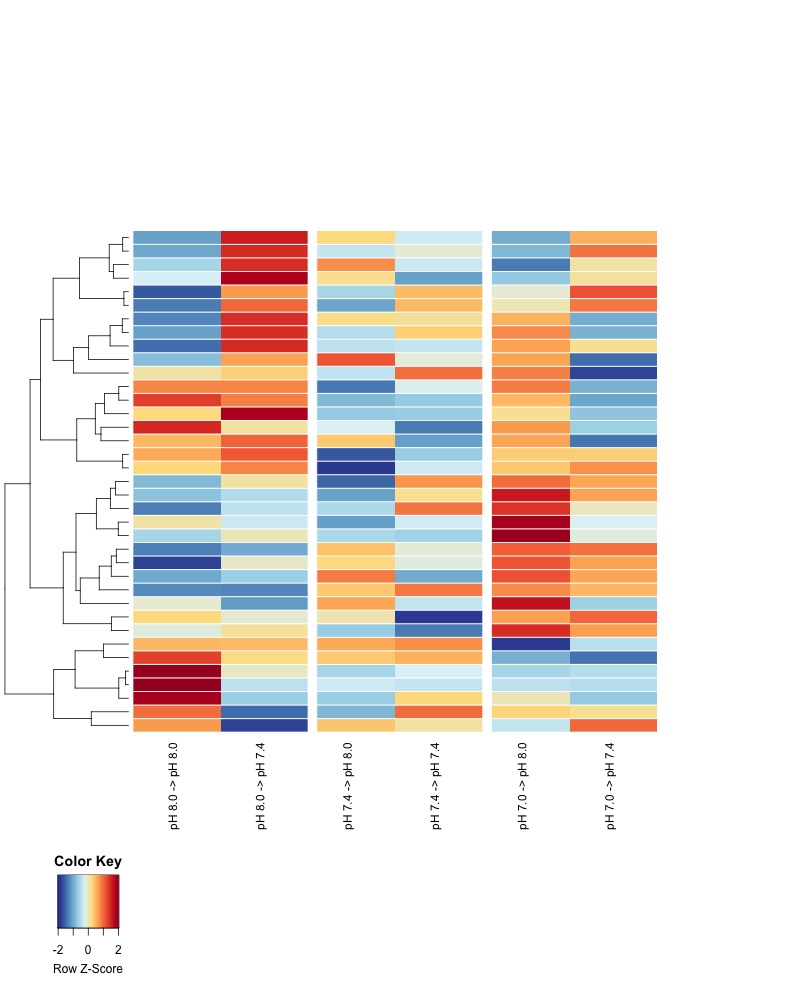
- Day 145 all pH samples:
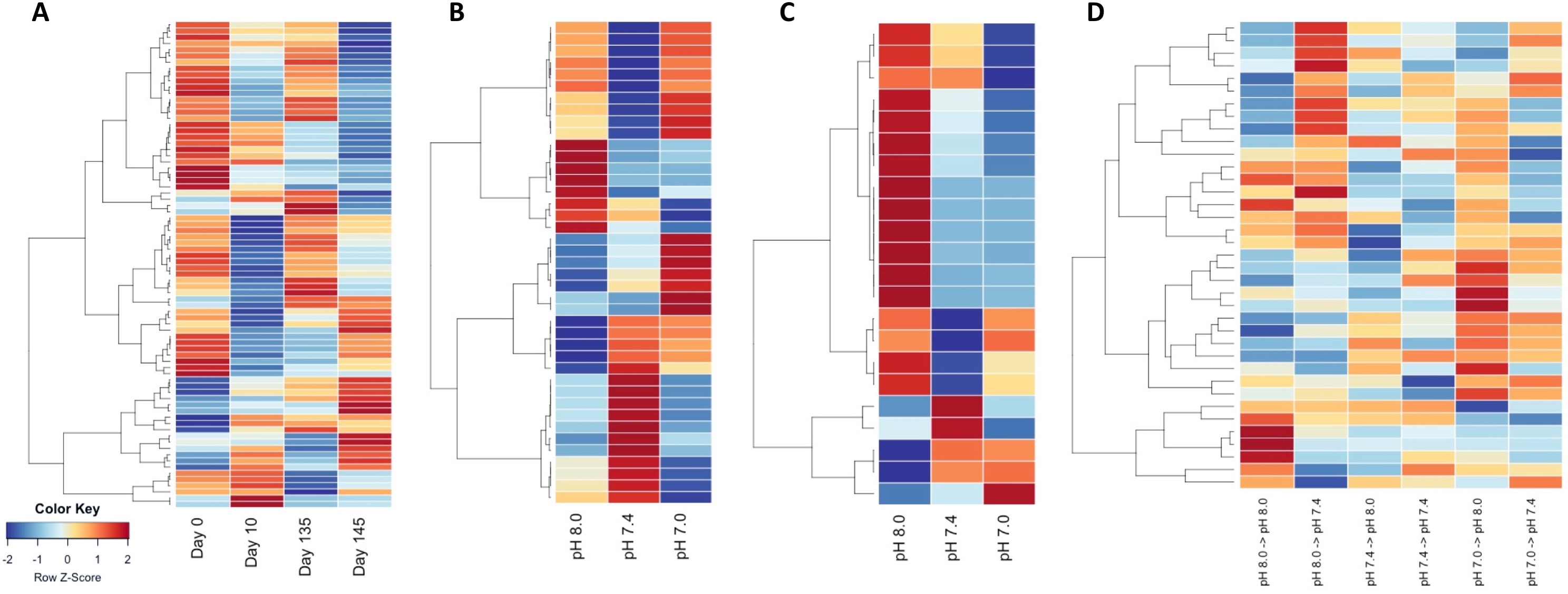
Filter DMRs, Run ANOVA on DMRs, Update heatmaps
- R project here: 20200320_anno.Rproj
- R markdown here: DMR_stats_and_anno.Rmd
- DMRs filtered for coverage in 3/4 samples per treatment group: -amb_AllTimes_DMR250bp_cov5x_rms_results_filtered.tsv
- DMRs significant at ANOVA p.val 0.05:
- Heatmaps of significant DMRs
- Other plots like data distribution before and after arcsin sqrt transformation and percent methylation plots facetted by individual DMRs can be found here: https://github.com/shellywanamaker/Shelly_Pgenerosa/blob/master/analyses/20200320_anno/
Next steps
- perform genomic feature analysis on DMRs vs background regions