This entry refers to IdentifyAndRemoveInconsistentProteins.md; R markdown file here.
I categorized proteins as being inconsistently detected between technical replicates if their adjusted normalized spectral abundance factor (ADJNSAF) values showed a standard deviation > 5 and a percent error > 20. This would remove a protein for instance if it was detected in one replicate at 100 and 0 in the other.
In total, this filter removed 3,396 proteins leaving 4,596 proteins to remain.
I then redid the PCA on these 4,596 proteins:
| filtered proteins | not filtered |
|---|---|
 |
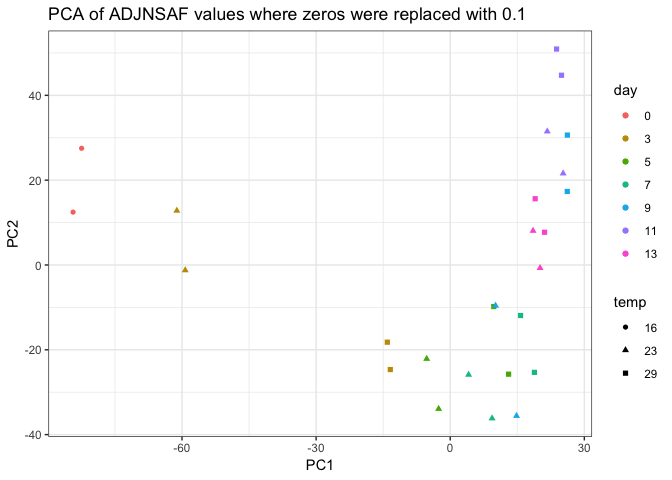 |
| filtered proteins | not filtered log vals |
|---|---|
 |
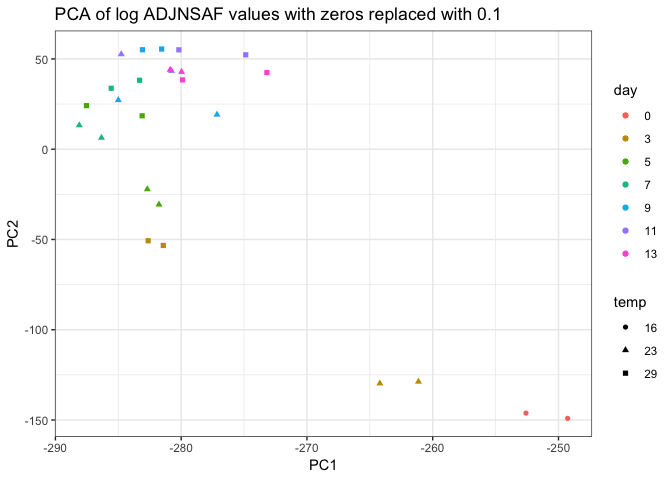 |
And redid the NMDS analyses:
| filtered proteins | not filtered |
|---|---|
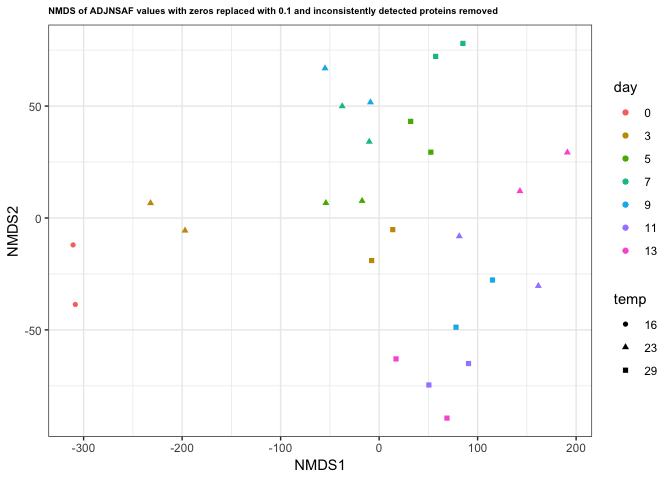 |
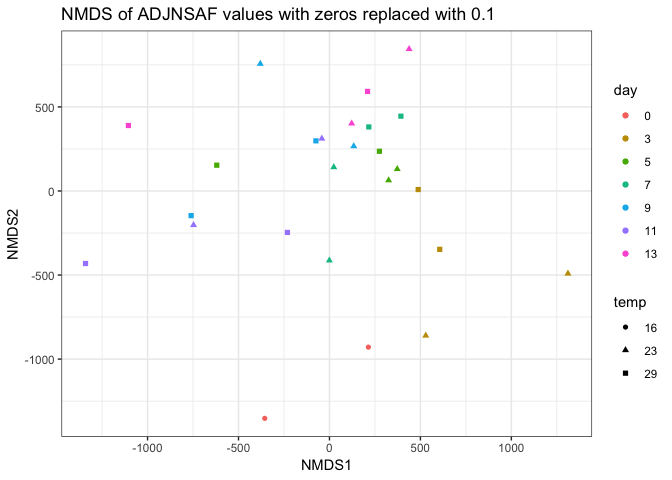 |
| filtered proteins | not filtered log vals |
|---|---|
 |
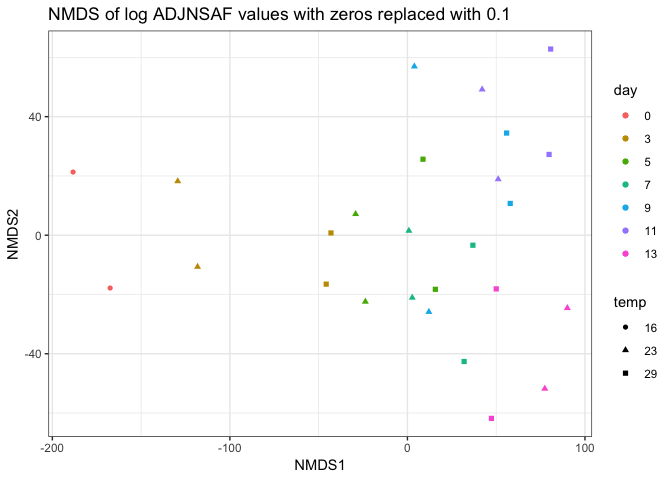 |
I think the NMDS on the filtered proteins shows the most agreement between technical replicates, but I’m interested to see what others think.
For now I went ahead with this filtered data set, averaged the NSAF values for technical replicates, and will proceed using that in downstream analyses. The filtered dataset with averaged technical replicate NSAF values called ‘silo3and9_nozerovals_noincnstprot.csv’ [can be found here] (https://github.com/shellywanamaker/OysterSeedProject/blob/master/analysis/nmds_R/silo3and9_nozerovals_noincnstprot.csv).