This entry is about trimming for the 2016 juvenile geoduck RRBS data Hollie generated.
Multi-core TrimGalore!
TrimGalore! can be run with multi-core settings if you use version 0.6.1 or newer. Reference to the multi-core TrimGalore! update is here: https://github.com/FelixKrueger/TrimGalore/pull/39. Using 8 cores reduced the run time for 100M reads from ~2hr10min to ~30min.
- script using multiple cores: 20200316_TG_speedTest.sh.
- For time differences, check these slurm.out files:
- multiple cores
- single core
- The single core slurm.out file was generated by this script: 20200306_TrimGfroger.sh.
Trimming history of RRBS data
- https://github.com/RobertsLab/resources/issues/812
- Initial trimming:
- 5/16/18: reads were trimmed with –rrbs –paired (default –directional and –illumina)
- Sam’s lab notebook
- Sam’s Jupyter Notebook
- sample EPI-167 Post-trimming fastqc = EPI-167 R1 EPI-167 R2
- sample EPI-167 MBias plots EPI-167
- 9/23/19: trimmed reads were further trimmed with a 5’ 20bp hard clip
- 5/16/18: reads were trimmed with –rrbs –paired (default –directional and –illumina)
Illumina Recommended Trimming:
- I spoke with Dina from Illumina tech support today and she found trimming recommendations for the Illumina Truseq Methylation Kit here on page 45: Illumina adapter sequences reference
- R1 Adapter: AGATCGGAAGAGCACACGTCTGAAC
- R2 Adapter: AGATCGGAAGAGCGTCGTGTAGGGA
- The first 13 bases (bolded above) correspond to the universal illumina adapter sequence you can specify in TrimGalore (AGATCGGAAGAGC)
- There is an additional 12bp added on by the Illumina Truseq Methylation Kit that are recommeneded to be trimmed off
- Bismark User Guide sections TruSeq DNA-Methylation Kit (formerly EpiGnome) and Random priming and 3’ Trimming in general
- Illumina Truseq Methylation Kit workflow
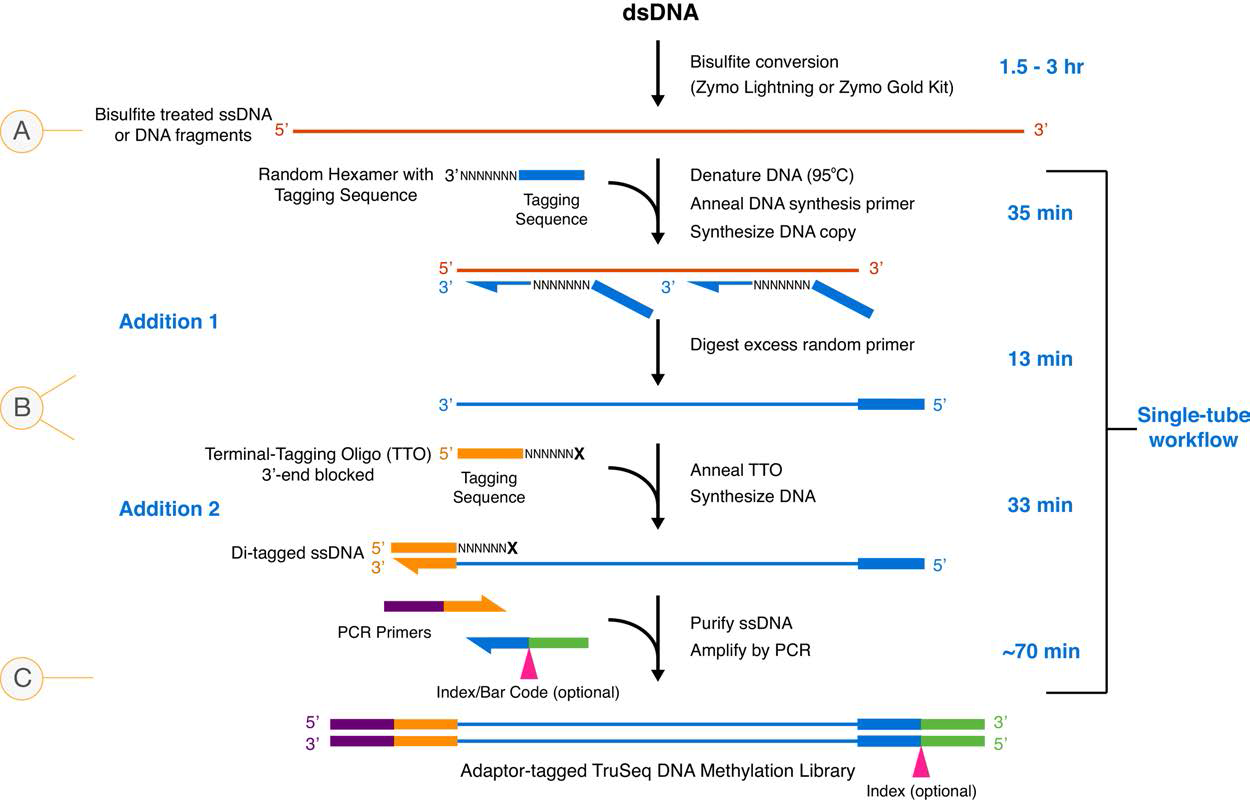
Testing Recommended Trimming Parameters
TrimGalore! with new parameters
I performed a test on just one sample: EPI-167
- Copied data to mox from nightingales:
(base) [strigg@mox2 raw]$ wget --no-check-certificate https://owl.fish.washington.edu/nightingales/P_generosa/EPI-167_S10_L002_R1_001.fastq.gz
--2020-03-16 20:29:13-- https://owl.fish.washington.edu/nightingales/P_generosa/EPI-167_S10_L002_R1_001.fastq.gz
Resolving owl.fish.washington.edu (owl.fish.washington.edu)... 128.95.149.83
Connecting to owl.fish.washington.edu (owl.fish.washington.edu)|128.95.149.83|:443... connected.
WARNING: cannot verify owl.fish.washington.edu's certificate, issued by ‘/C=US/ST=MI/L=Ann Arbor/O=Internet2/OU=InCommon/CN=InCommon RSA Server CA’:
Unable to locally verify the issuer's authority.
HTTP request sent, awaiting response... 200 OK
Length: 1451174652 (1.4G) [application/x-gzip]
Saving to: ‘EPI-167_S10_L002_R1_001.fastq.gz’
100%[=============================================================================================>] 1,451,174,652 27.6MB/s in 51s
2020-03-16 20:30:04 (26.9 MB/s) - ‘EPI-167_S10_L002_R1_001.fastq.gz’ saved [1451174652/1451174652]
(base) [strigg@mox2 raw]$ wget --no-check-certificate https://owl.fish.washington.edu/nightingales/P_generosa/EPI-167_S10_L002_R2_001.fastq.gz
--2020-03-16 20:30:08-- https://owl.fish.washington.edu/nightingales/P_generosa/EPI-167_S10_L002_R2_001.fastq.gz
Resolving owl.fish.washington.edu (owl.fish.washington.edu)... 128.95.149.83
Connecting to owl.fish.washington.edu (owl.fish.washington.edu)|128.95.149.83|:443... connected.
WARNING: cannot verify owl.fish.washington.edu's certificate, issued by ‘/C=US/ST=MI/L=Ann Arbor/O=Internet2/OU=InCommon/CN=InCommon RSA Server CA’:
Unable to locally verify the issuer's authority.
HTTP request sent, awaiting response... 200 OK
Length: 1496018906 (1.4G) [application/x-gzip]
Saving to: ‘EPI-167_S10_L002_R2_001.fastq.gz’
100%[=============================================================================================>] 1,496,018,906 27.2MB/s in 53s
2020-03-16 20:31:01 (27.1 MB/s) - ‘EPI-167_S10_L002_R2_001.fastq.gz’ saved [1496018906/1496018906]
- Trim the 25bp adapter sequences:
- script 20200316_TG_EPI-Test.sh
- slurm file: slurm-2268256.out
- output: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200316/TG_EPI-Test1/
- fastqc R1: EPI-167_S10_L002_R1_001_val_1_fastqc.html
- fastqc R2: EPI-167_S10_L002_R2_001_val_2_fastqc.html
- CONCLUSION: The reads don’t look any better than they did in the original trimming Sam did:
- Trim with 5’ 25bp clip and a 3’ 10bp clip and trim the 25bp adapter sequences:
- script 20200316_TG_EPI-Test2.sh
- slurm file: slurm-2268263.out
- output: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20200316/TG_EPI-Test2/
- fastqc R1: EPI-167_S10_L002_R1_001_val_1_fastqc.html
- fastqc R2: EPI-167_S10_L002_R2_001_val_2_fastqc.html
- CONCLUSIONS: The reads look improved, but are now only ~65bp long
Alignments with new trimming
- ran this script 20200316_BmrkAln_EpiTest2.sh
- NEXT STEPS:
- check Mbias plots in report
- check percent methylation
- previous
-
Trim date 03/16/2020 05/16/2018 Read pairs analyzed 23436512 24481250 mapping efficiency (%) 40.9 42.6 ambiguously mapped read pairs (%) 11.8 8.2 unaligned read pairs 47.3 49.2 mC in CpG (%) 25.3 27.9 mC in CHG (%) 1.7 2.9 mC in CHH (%) 2.7 3 mC in CN or CHN (%) 4.9 8.5
- determine if deduplicating should be done
- previous report showed 26.85% duplicate alignments were removed - NOTE: previous alignments were done using genome v074. Although there shouldn’t be a difference between this genome and the one on OFS (Panopea-generosa-v1.0.fa), I am currently performing alignment of the 5/16/19 trimmed reads and the 9/23/19 trimmed reads for EPI-167/