Purpose
To see how variants called from Biscuit (WGBS) and rnavar (RNAseq) pipelines compare.
Results
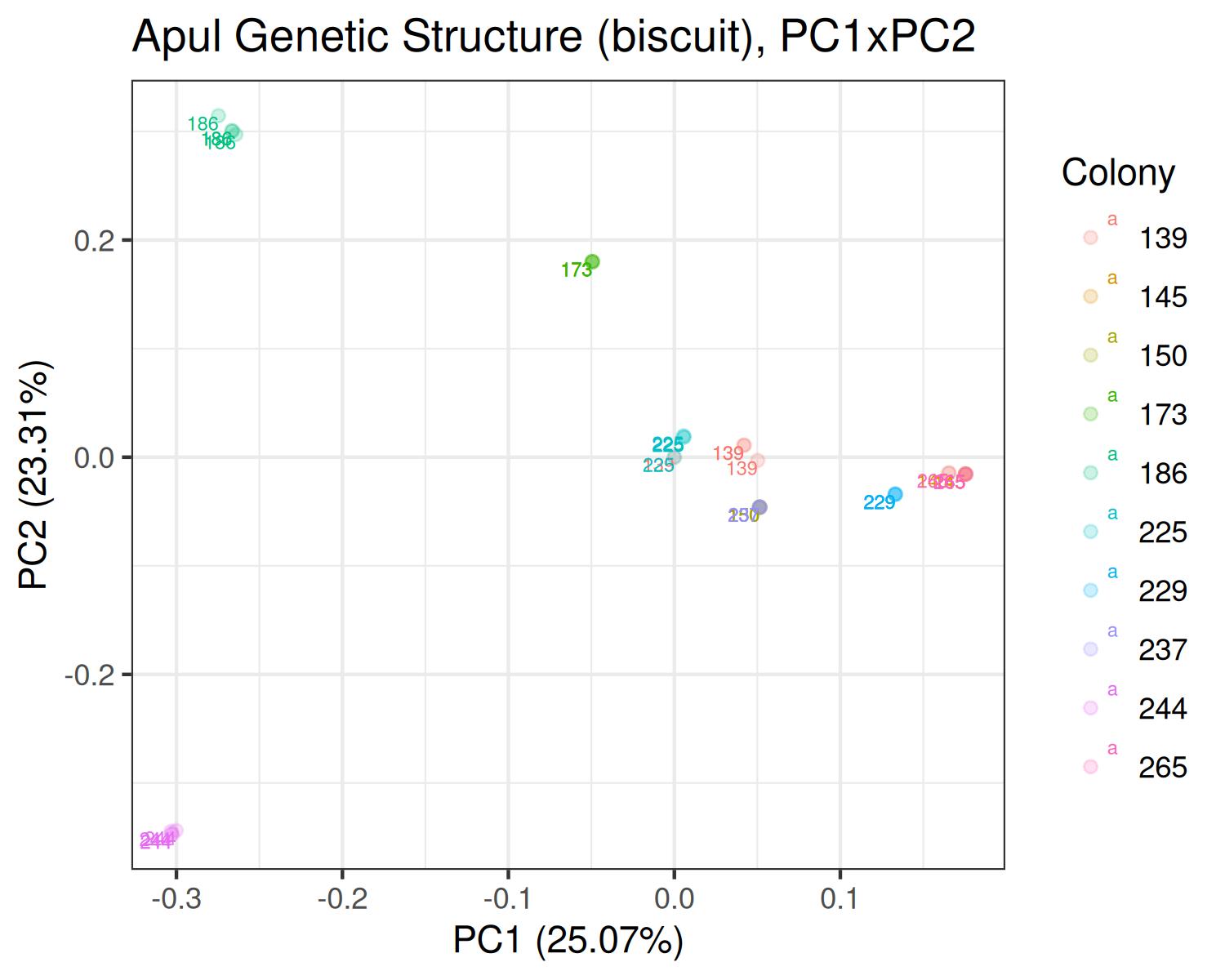
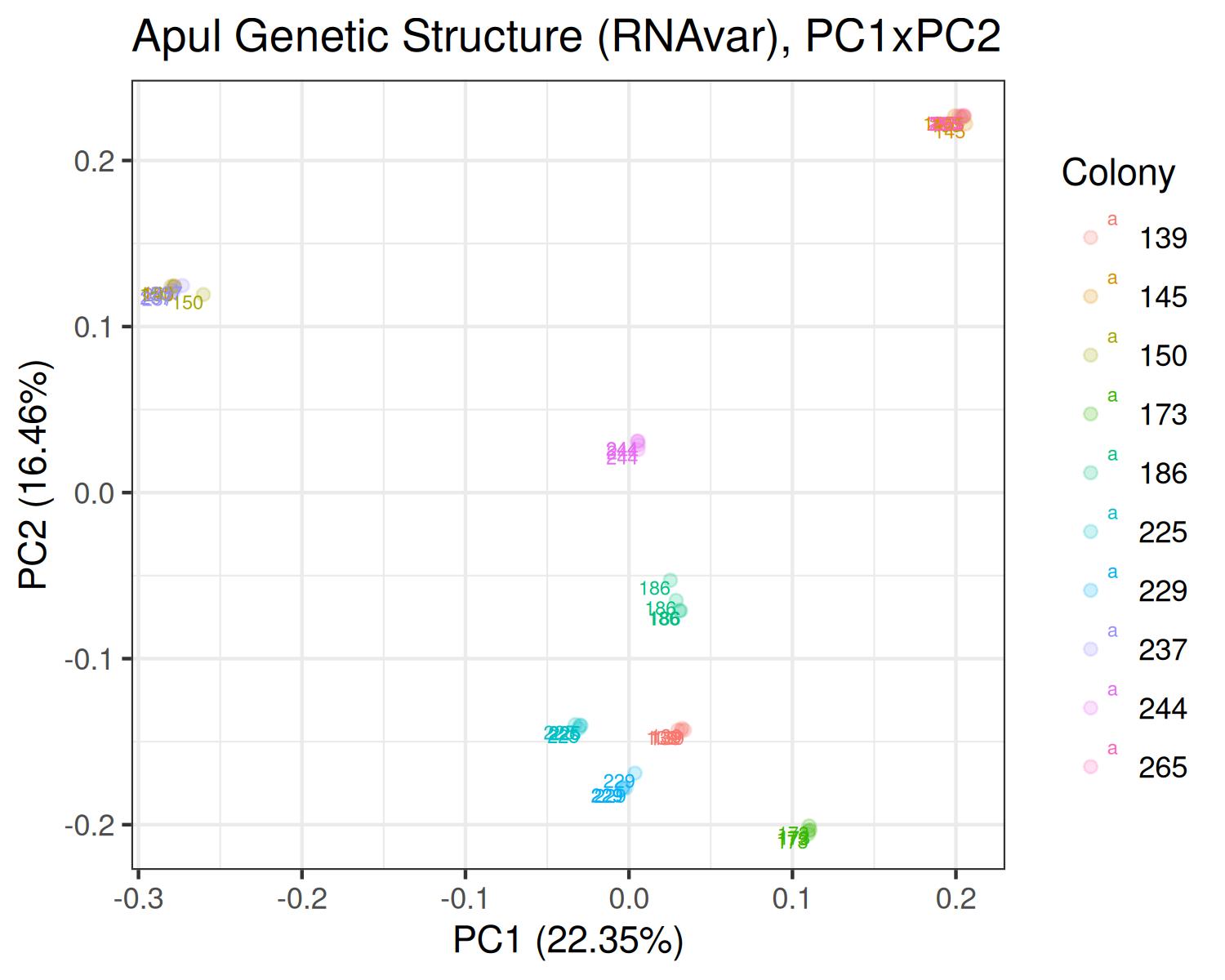
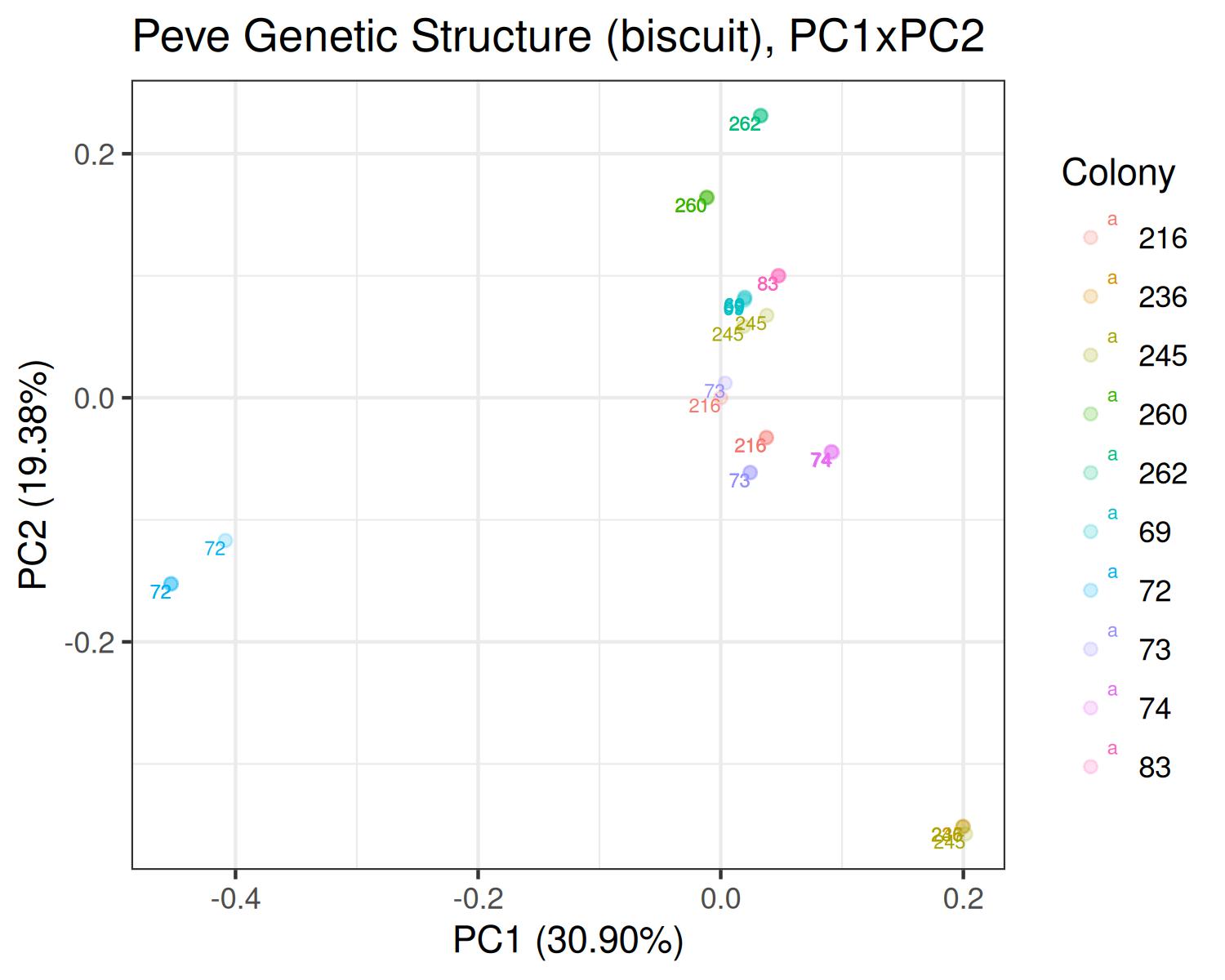
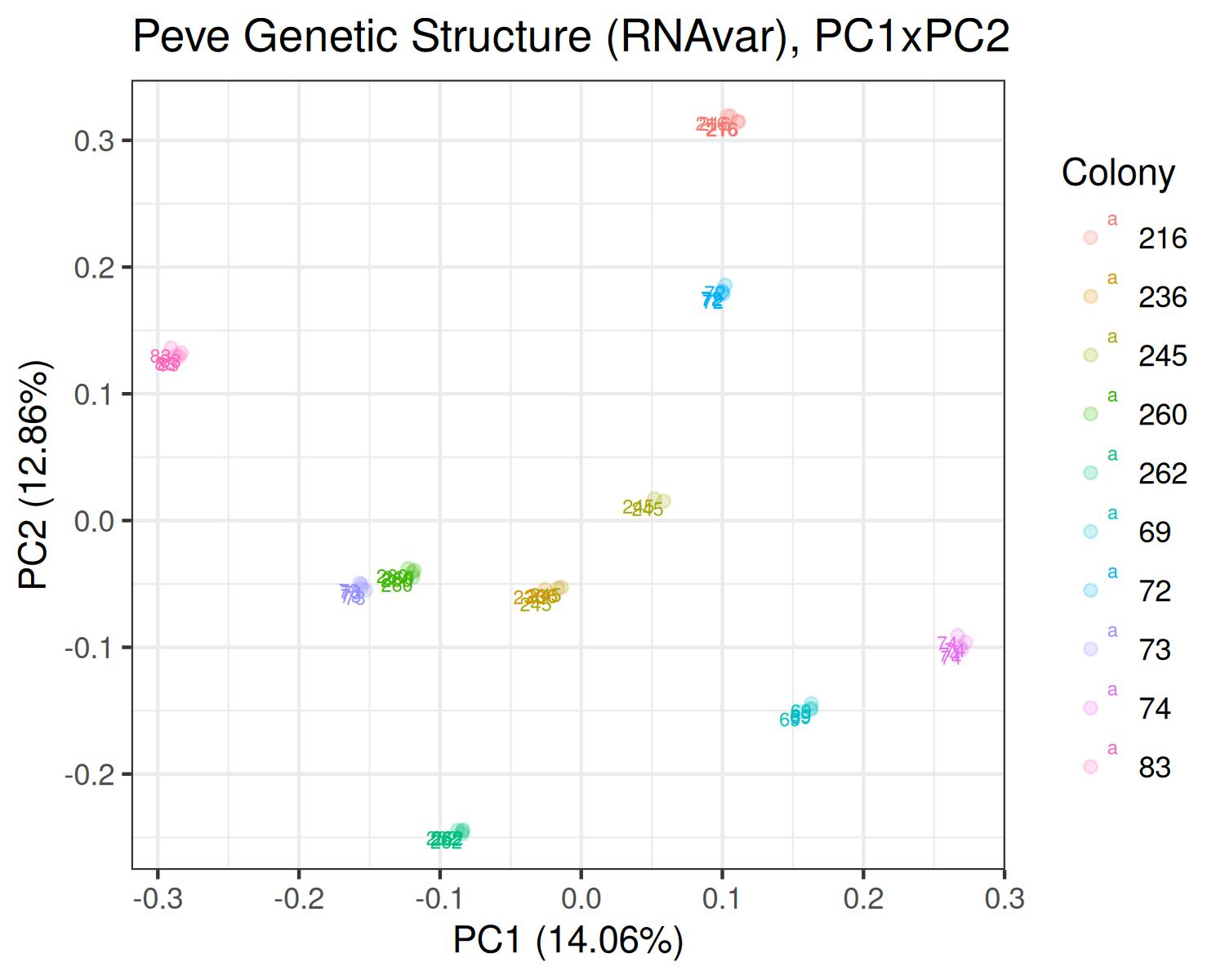
PCA
Apul
- Variants called from WGBS:
- Variants called from RNAseq:
Peve
- Variants called from WGBS:
- Variants called from RNAseq:
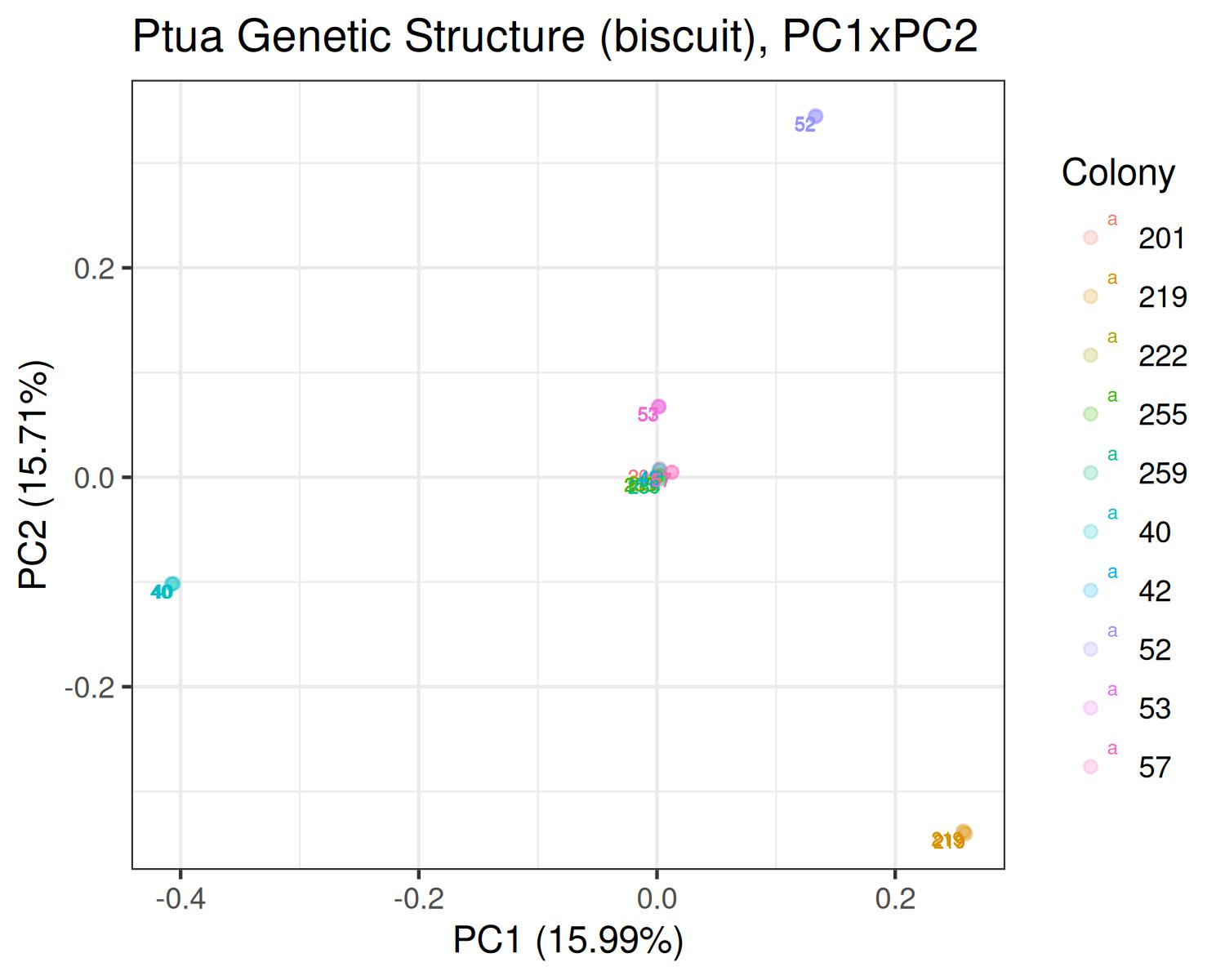
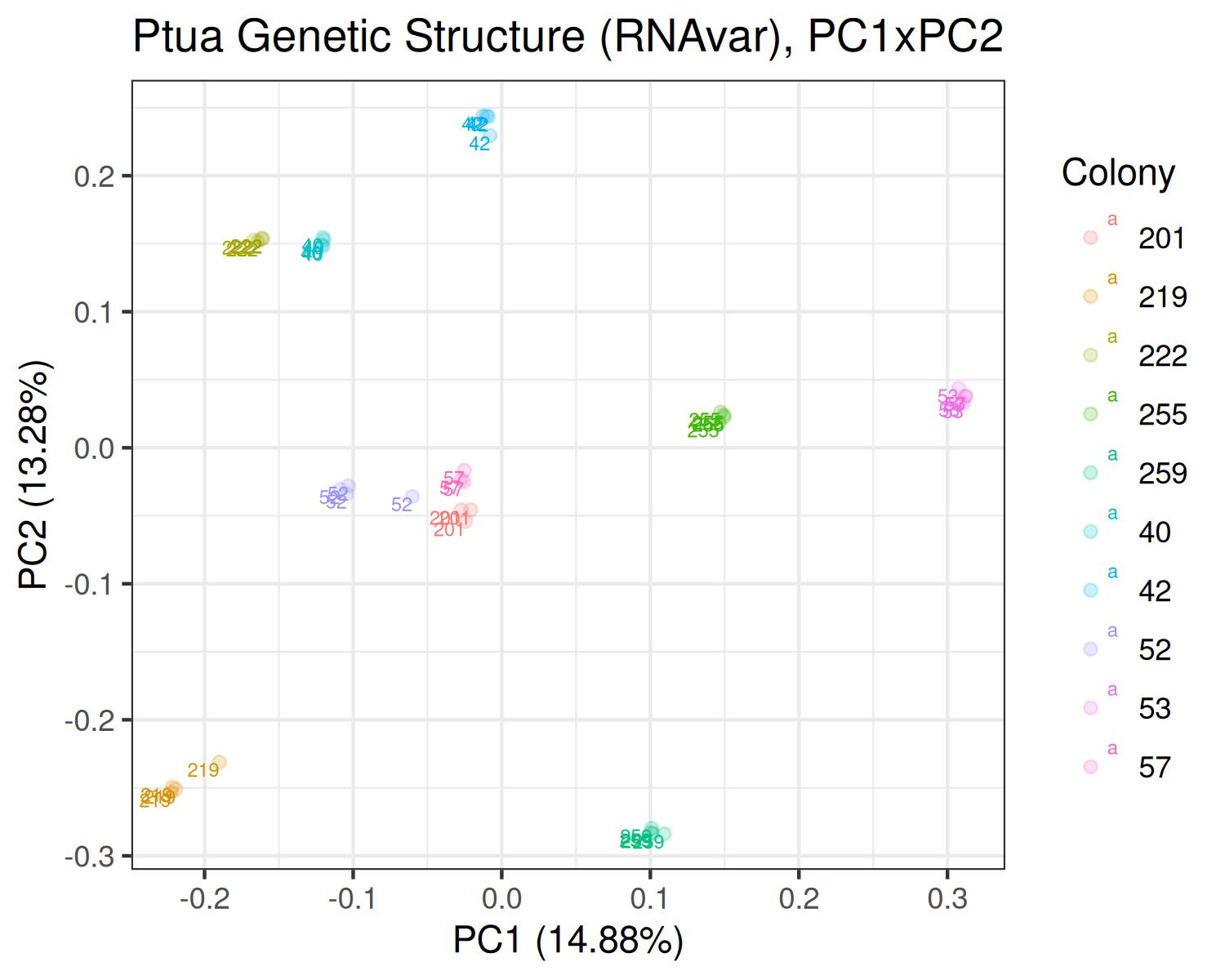
Ptua
- Variants called from WGBS:
- Variants called from RNAseq:
Number of loci
Apul
- Biscuit all filtered: 171
- rnavar all filtered: 1,173
- Biscuit 1st filter: 124,899
- rnavar 1st filter: 75,677
- Biscuit all variants:
- rnavar all variants: 986,105
Peve
- Biscuit all filtered: 184
- rnavar all filtered: 1,028
- Biscuit 1st filter: 113,700
- rnavar 1st filter: 54,352
- Biscuit all variants:
- rnavar all variants: 931,363
Ptua
- Biscuit all filtered: 208,959
- rnavar all filtered: 1,934
- Biscuit 1st filter:
- rnavar 1st filter: 127,588
- Biscuit all variants:
- rnavar all variants:
Methods
PCA
- 04_Genetic-Structure-Analysis.Rmd. This code was modified from Laura Spencer’s red king crab SNP analysis here: https://github.com/laurahspencer/red-king_RNASeq-2022/blob/main/notebooks/04_Genetic-Structure-Analysis.Rmd
source data files used in the code can be found here in subdirectories: https://gannet.fish.washington.edu/metacarcinus/E5/. These files are the output from the analyses done here:
- Call variants from Apul, Peve, and Ptua RNA-seq data
- Call variants from Apul and Peve WGBS data
- Identify variants in RNAseq and EM-seq data from P.tuahiniensis
Number of loci
awk '{if($0!~/#/)print $0}' Peve.rnavar.merged-filtered-true-all.vcf | wc -l