Sample details
- 2 lice samples from 20190523
- Samples from 20191118 (in fridge in 209)
- 2 pools of adult sea lice from different populations
- from two different salmon farms
- different salinities? temperatures?
- *** need more info about both of sample groups; waiting to hear back from Cristian’s lab
Methylation data
re-trimmed all samples with bismark recommended trimming parameters
aligned trimmed reads to genome
| desc. stat |
Sealice F1 S20 |
Sealice F2 S22 |
| total reads before trim |
226181237 |
163876559 |
| perc reads trim removed |
0.56 |
0.43 |
| total reads after trim |
224907116 |
163179589 |
| uniq aligned reads |
62174415 |
34864893 |
| perc uniq aligned |
27.64 |
21.37 |
| ambig reads |
49967995 |
31414190 |
| perc ambig aligned |
22.22 |
19.25 |
| perc no align |
50.14 |
59.38 |
| dedup reads |
39280061 |
26425601 |
| dedup reads percent |
63.18 |
75.79 |
| dup reads |
22894354 |
8439292 |
| dup reads percent |
36.82 |
24.21 |
| percent cpg meth |
1.1 |
1.0 |
| percent chg meth |
1.0 |
0.8 |
| percent chh meth |
1.4 |
1.5 |
- qualimap summary of alignments:
prepared merged CpG 5x cov files
categorized CpGs with 5x cov:
5x CpG summary tables:
Sample|Methylated CpG (>= 50%)|Sparsely methylated CpG (10% - 50%)|Unmethylated CpG (< 10%)
:—–:|:—–:|:—–:|:—–:
F1|2335|391515|8890795
F2|1864|2232274|6108325
CpG category|F1:F2 CpG overlap|Uniq F1 CpGs|Uniq F2 CpGs|frac F1 mCpG overlapping|frac F2 mCpG overlapping
:—–:|:—–:|:—–:|:—–:|:—–:|:—–:
Methylated CpG(>= 50%) | 545 | 1790 | 1319 |23.34 | 29.24
Sparsely methylated CpG(10% - 50%) | 19838 | 371677 |212436| 5.07 | 8.54
Unmethylated CpG(< 10%) |5431008 |3459787| 677317 |61.09| 88.91
5x merged CpG summary tables:
| Sample |
Methylated CpG (>= 50%) |
Sparsely methylated CpG (10% - 50%) |
Unmethylated CpG (< 10%) |
| F1 |
1342 |
233941 |
6831337 |
| F2 |
1102 |
165423 |
5130433 |
| CpG category |
F1:F2 CpG overlap |
Uniq F1 CpGs |
Uniq F2 CpGs |
frac F1 mCpG overlapping |
frac F2 mCpG overlapping |
| Methylated CpG(>= 50%) |
314 |
1028 |
788 |
23.40 |
28.49 |
| Sparsely methylated CpG(10% - 50%) |
13570 |
220371 |
151853 |
5.80 |
8.20 |
| Unmethylated CpG(< 10%) |
4824175 |
2007162 |
306258 |
70.62 |
94.03 |
IGV session
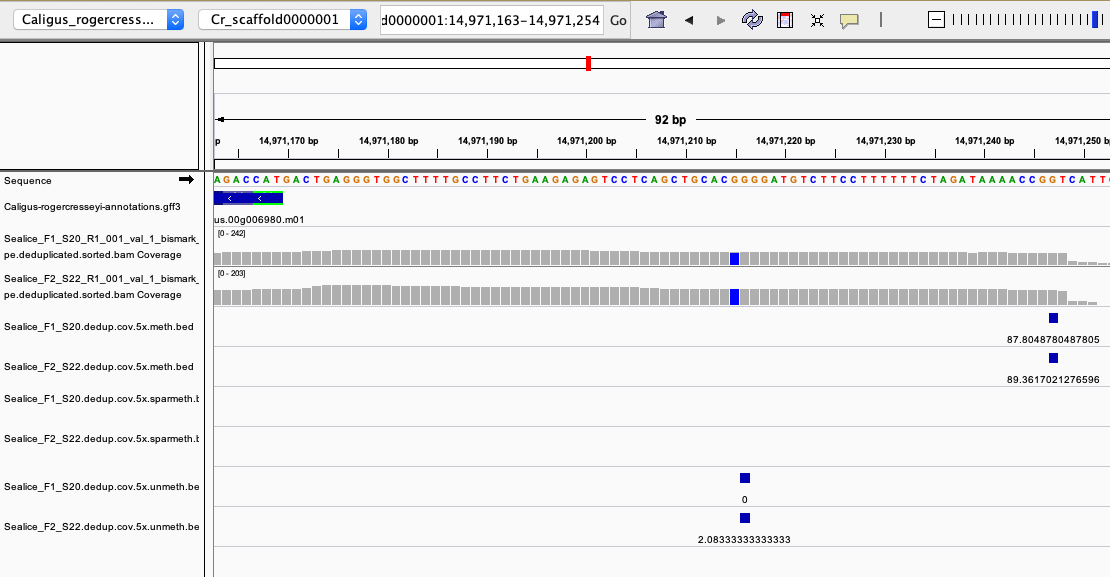
Example of highly methylated CpG overlapping between both samples
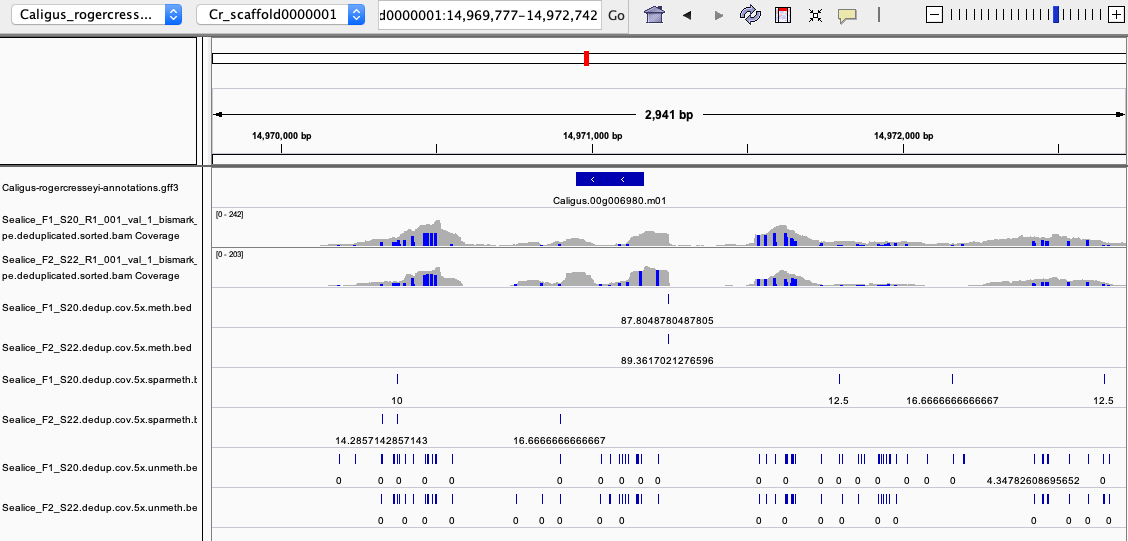
zoomed in view
Genomic Feature analysis
| Feature |
num. of features |
| CDS |
30022 |
| exon |
30022 |
| gene |
23686 |
| mRNA |
23686 |
- Checked for features overlapping with CpGs methylated >=50%
| Sample |
mCpG(>= 50%) |
mCpG overlapping with genes/mRNA |
mCpG overlapping with exon/CDS |
| F1 |
2335 |
114 |
106 |
| F2 |
1864 |
103 |
95 |
| F1.merged |
1342 |
60 |
58 |
| F2.merged |
1102 |
45 |
42 |
Moving forward
- Overall most mCpGs are not located in genic regions so it’s hard to say how to target this sparse methylation aside from MBD
- Would be great to get repeat regions and other features (UTR, etc)
- Options for 1 sequencing run:
- resequence 2 individuals to attempt to acheive 100% genome coverage
- this would give at least 500M reads more data per individual
- cost: $4,940 (1 Novaseq run)
- WGBS 4 individuals aiming for 400M reads each to attempt acheive 5x coverage of >95% of genome
- cost: ~$5150 = $4,940 (1 Novaseq run) + library prep (~ $50/sample) + time
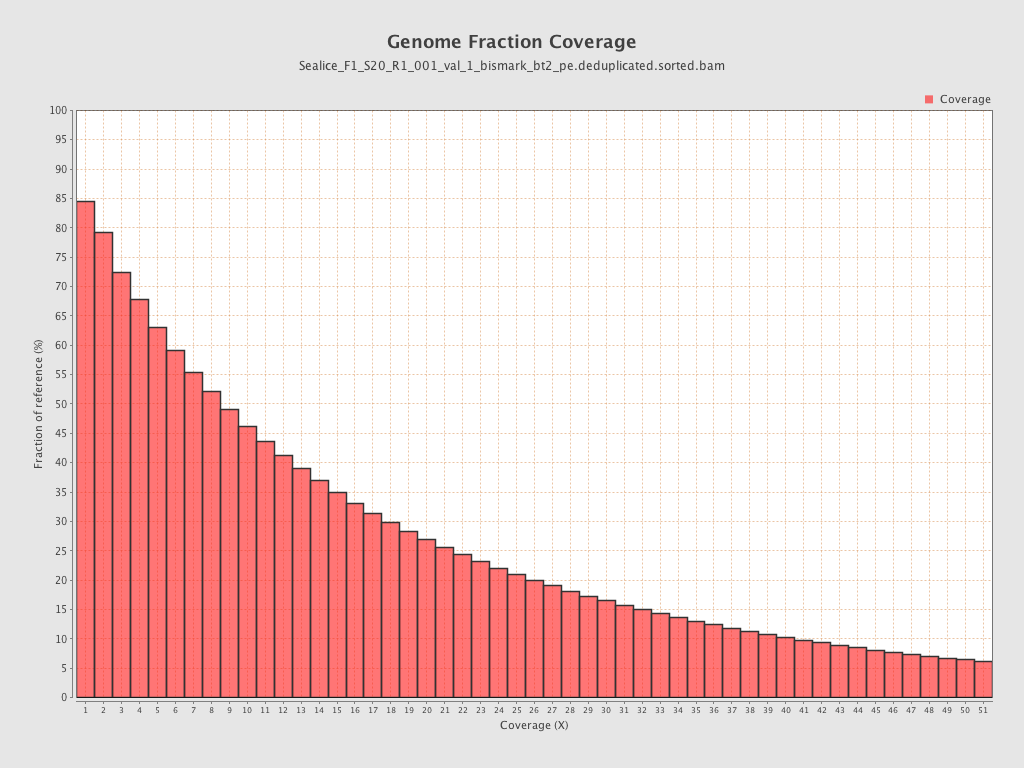
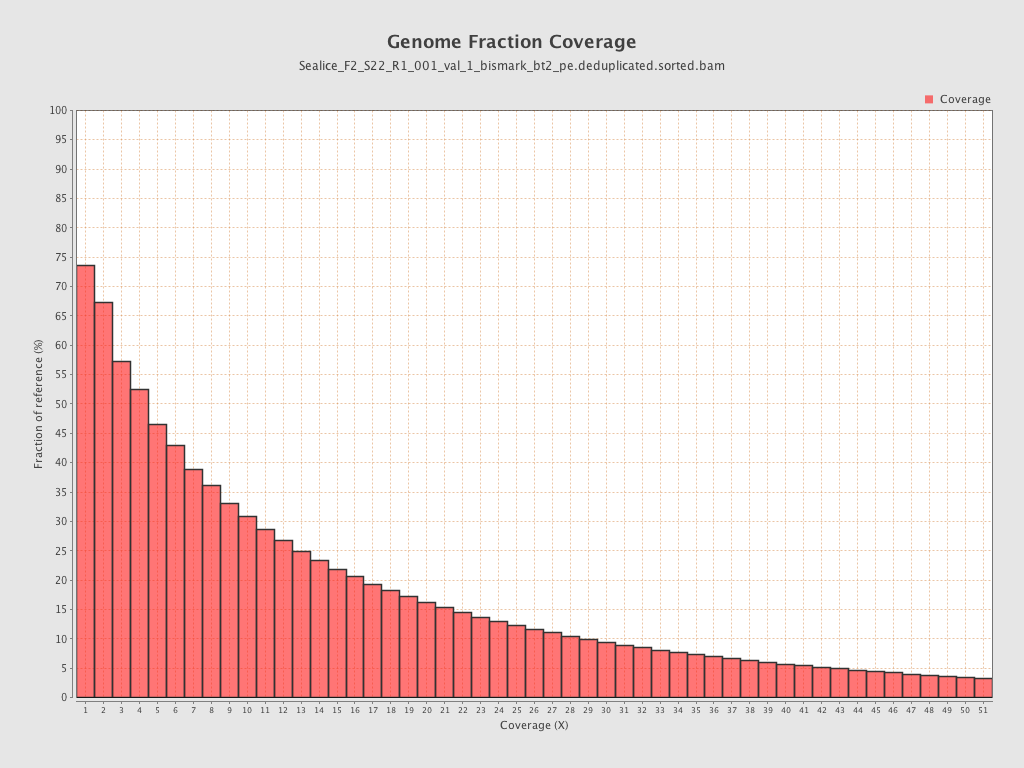
- logic: 226M reads gave 65% genome coverage @5x read depth, 168M gave 45% genome coverage @5x read depth
- assuming a linear relationship between read depth and genome coverage (2.9 * 100) + 37.5 = ~340M ; see chart below
.png)
- WGBS 2-3 individuals aiming for 500M reads each to attempt to achieve 10x coverage of > 95% of genome
- cost: ~$5100 = $4,940 (1 Novaseq run) + library prep (~ $50/sample) + time
- logic: 226M reads gave 46% genome coverage @ 10x read depth, 168M gave 31% genome coverage @10x read depth
- assuming a linear relationship between read depth and genome coverage (3.77 * 100) + 51.8 = ~430M ; see chart below
.png)
- MBD-BS on 10 individuals from each population:



.png)
.png)