This analysis is a follow up to Wed. Oct 23 analysis
Rerun DMRfind with different parameters
Reran DMRfind with different MCmax settings (this specifies what a differentially methylated site (DMS) is; it allows loci to not be exactly be overlapping but be within a window to be considered a DMS which helps for low coverage samples). This window size is defined by MCmax:
- mox scripts here for the following MCmax window sizes:
- 50bp (analysis from Oct 23): 20191023_DMRfindAllEPI.sh
- 30bp: 20191024_DMRfindAllEPI.sh
- 25bp: 20191024_DMRfind_allEPI_25bp.sh
- 10bp: 20191024_DMRfind_allEPI_10bp.sh
- output files here:
- 50bp: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20191023/
- 30bp: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20191024/
- 25bp: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20191024_25bp/
- 10bp: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20191024_10bp/
Validate DMR bed files in IGV
- loaded the following bed files into IGV:
- 50bp: amb_AllTimes_DMR250bp_MCmax50_cov5x_rms_results_collapsed.tsv.DMR.bed
- 30bp: amb_AllTimes_DMR250bp_MCmax30_cov5x_rms_results_collapsed.tsv.DMR.bed
- 25bp: amb_AllTimes_DMR250bp_MCmax25_cov5x_rms_results_collapsed.tsv.DMR.bed
- 10bp: amb_AllTimes_DMR250bp_MCmax10_cov5x_rms_results_collapsed.tsv.DMR.bed
- bam files: amb all times filtered bam files
- IGV session here: amb_AllTimes_IGV_20191024.xml
- Interestingly it seems there are some DMRs that make sense and are identified by all parameter settings like this example:
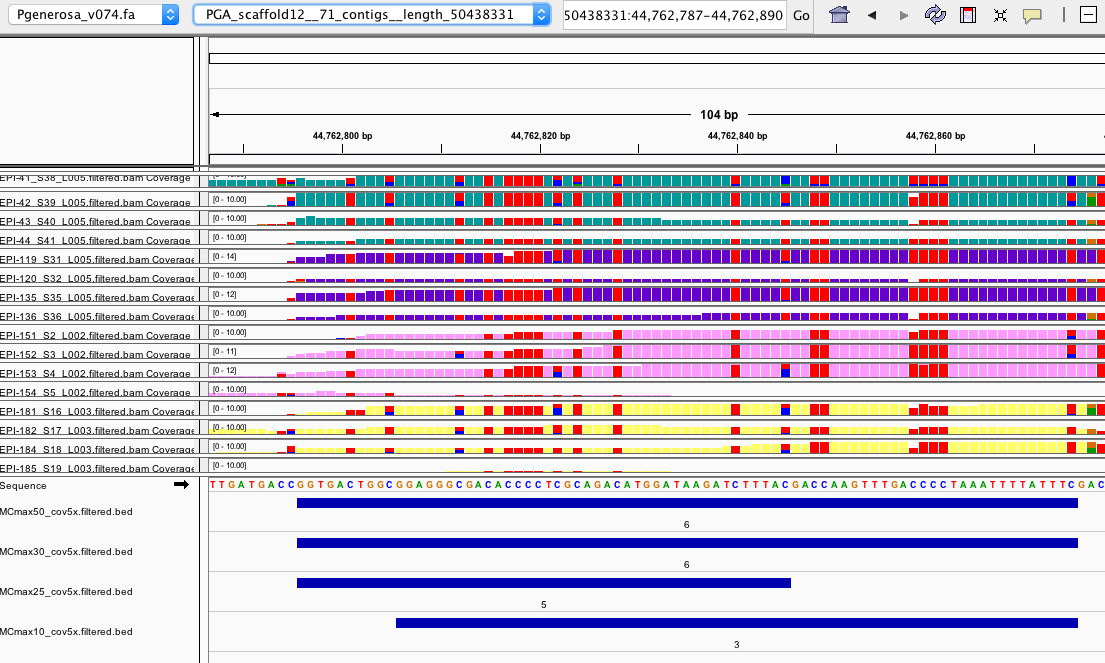 and
and 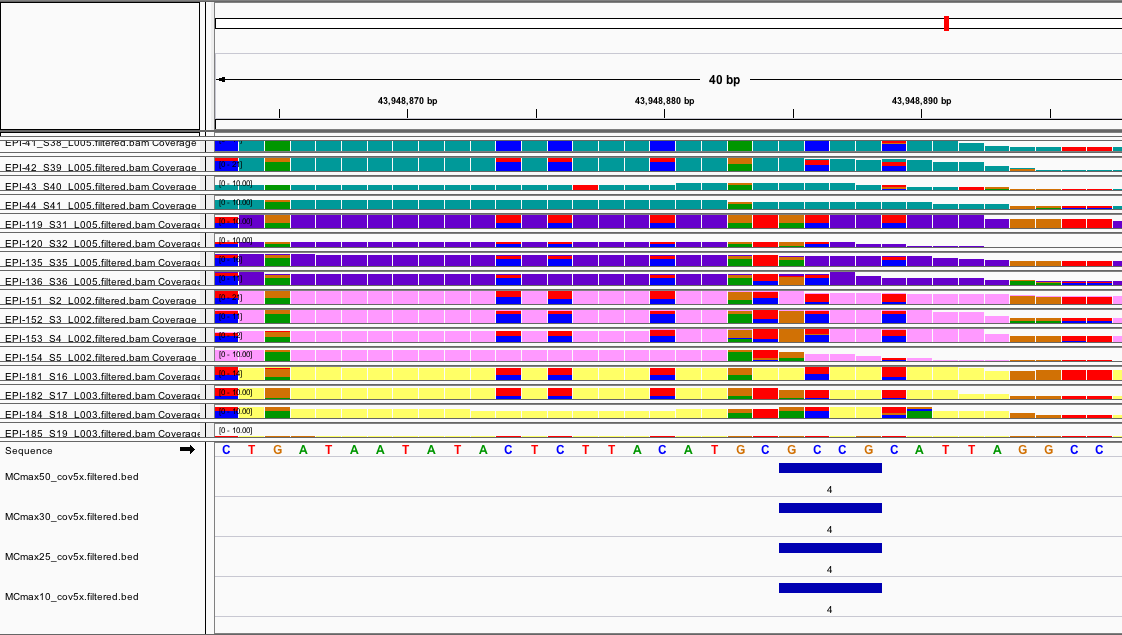
- Then there are some DMRs that make sense and are only identified by one parameter setting like this example:

- Then some DMRs that are only identified by one setting and don’t make sense like this example:
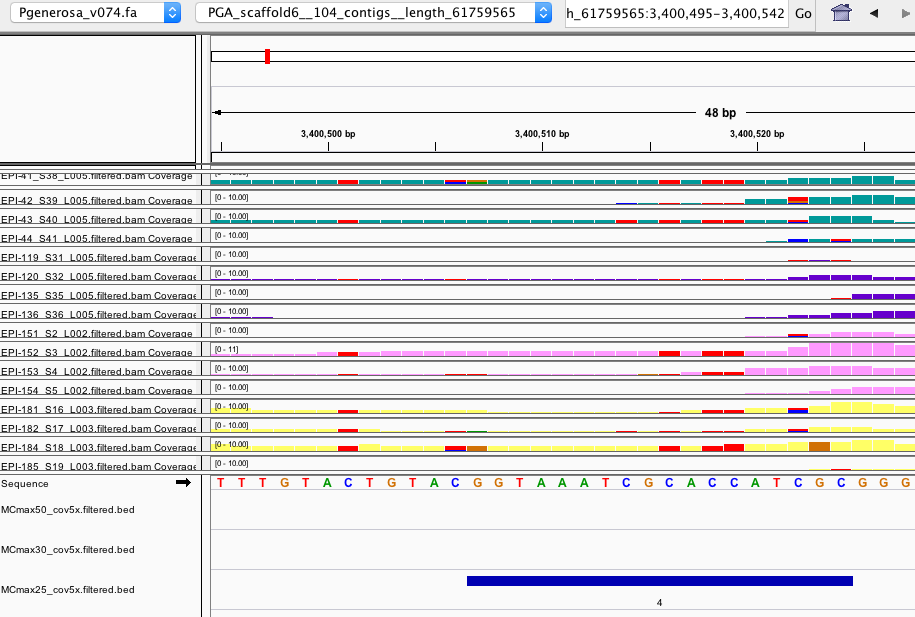
CONCLUSIONS
- Try running group stats on % methylation data and see if it excludes DMRs that don’t make sense
- Yupeng confirmed that methylpy only runs statistics on within sample data, not on group data. So I need to apply an ANOVA or GLM to determine DMRs that are statistically different between groups