Rerun DMRfind with looser parameters
Reran DMRfind using loose parameters to be able to filter down by coverage in 3 samples/category
- mox script here: 20191023_DMRfindAllEPI.sh
- DMRfind parameters:
- minimum coverage = 5 reads
- mc-max dist = 50 bp (this specifies what a differentially methylated site is; I set this to 50 bp so that sites can be overlapping within a 50bp window to be considered which helps for low coverage samples)
- dmr-max-dist = 250bp (this specifies the max size of the DMR)
- min-num-dms = 3 (this specifies the number of DMS within a DMR required for the DMR to be called)
- output files here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20191023/
Filter DMR output
- created this R project for filtering DMR output files for only DMRs with coverage in at least 3/4 samples per experimental catagory.
- R script here: DMR_cov_in_0.75_SamplesPerCategory.R
- filtered output files here: https://github.com/shellywanamaker/Shelly_Pgenerosa/tree/master/analyses/DMR_cov_in_0.75_SamplesPerCategory
- Used the filtered DMR output files above to filter DMR bed files using this jupyter notebook: 20191023_DMRfind_allEPI.ipynb
Validate filtered DMRs
- I only did this for all ambient sample comparison because I had previously made filtered bam files for doing this comparison (see oct 1 post)
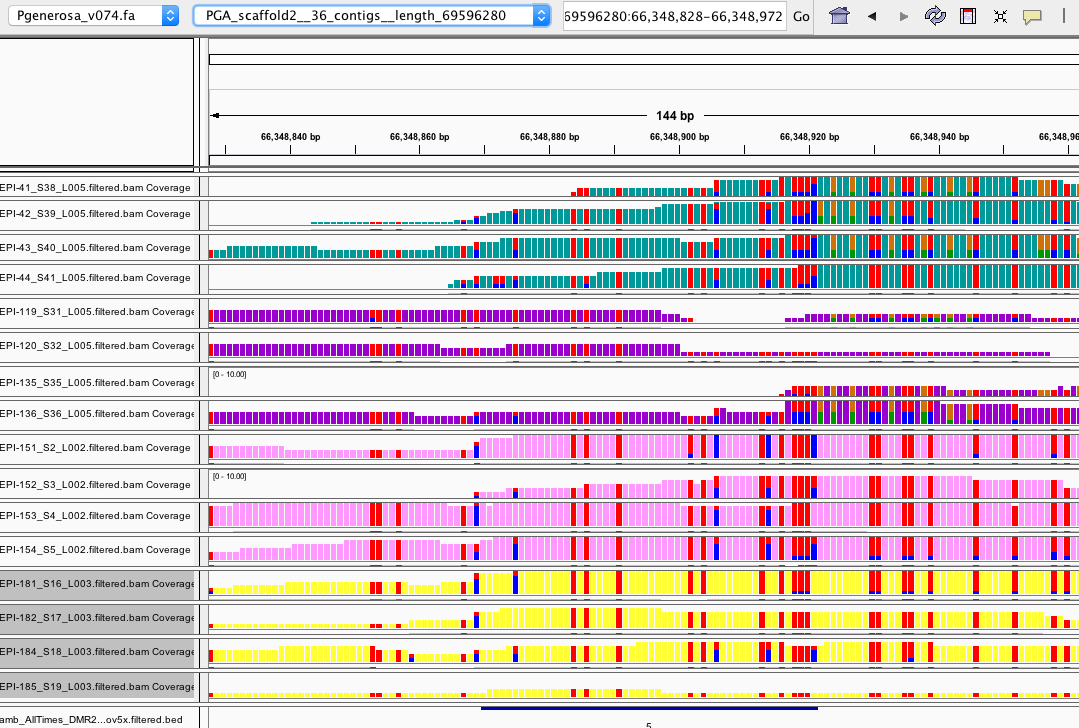
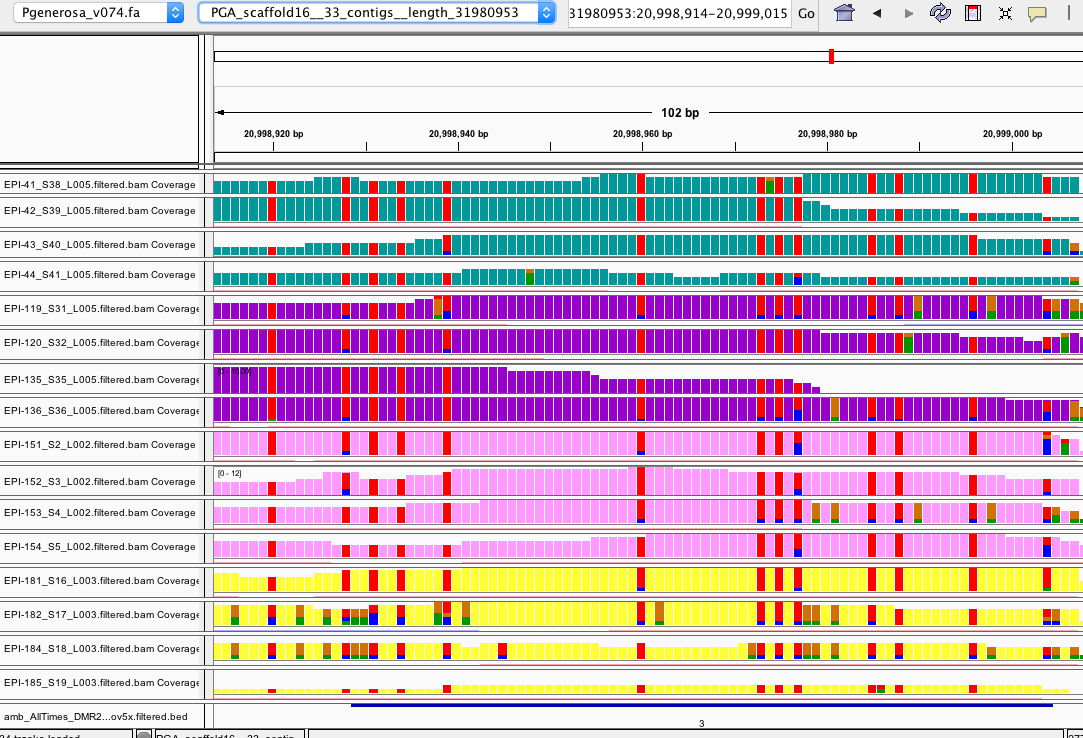
- looked at a number of DMRs for all ambient sample comparison in IGV
- IGV session here: amb_AllTimes_IGV.xml
- files uploaded:
- I think most make sense




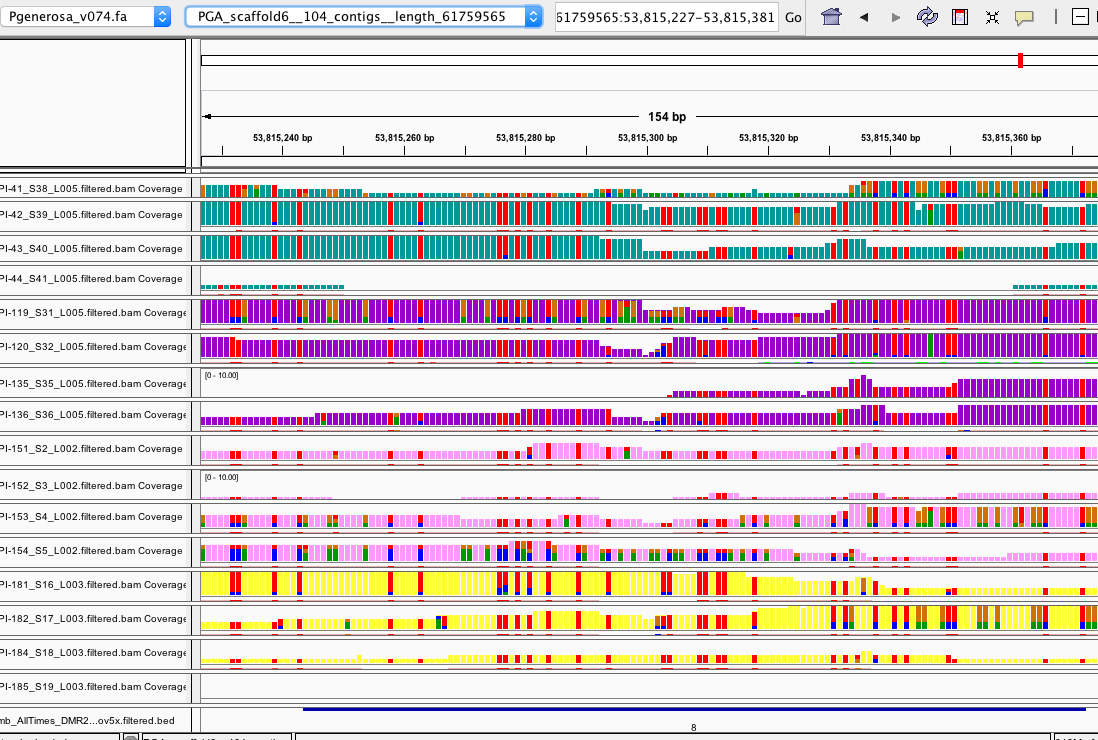
 some are less convincing:
some are less convincing:

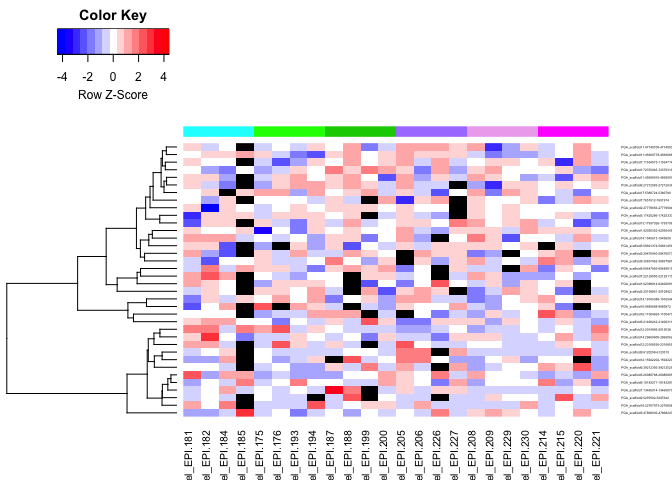
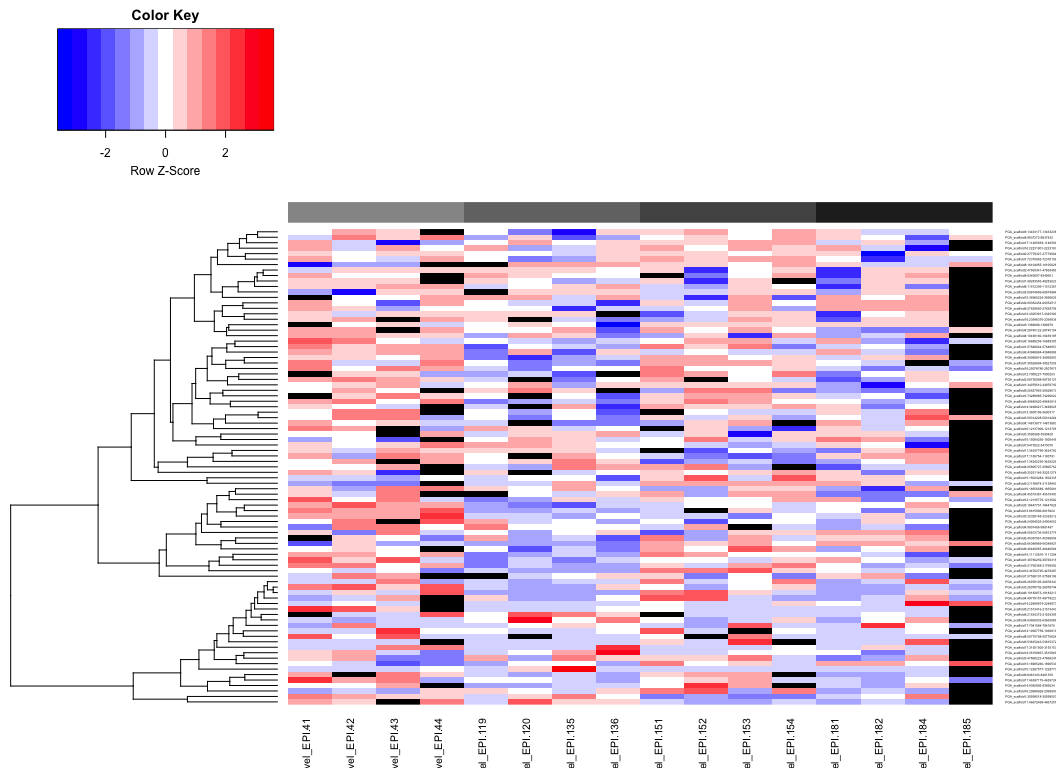
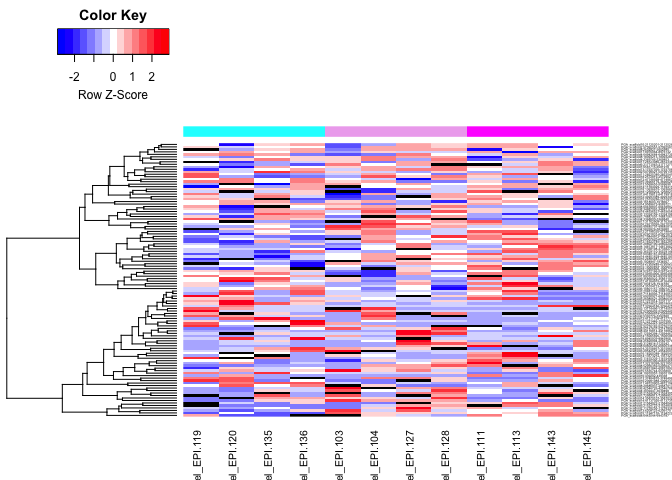
heatmaps of DMRs
- markdown file here: Oct23_DMR_heatmaps.md
- all ambient times (lighter - darker gray = Day 0 - Day 145):
- day 10 all pH (teal = ambient, light pink = low pH, dark pink = super low pH):
- day 135 all pH (teal = ambient, light pink = low pH, dark pink = super low pH):

- day 145 all pH (teal = ambient-ambient, light green = ambient-low pH, dark green = ambient-super low pH, purple = low-ambient, light pink = low-low, dark pink = low - superlow):