Network analysis on same-day temperature ChiSq pvalue 0.1 selected proteins
- made new R project ChiSqPvalproteins_GOsemsim_edges.Rproj and new cleaned up code CreateNodeAndGoSemSimEdgeAttributeFiles.R to generate network files based on proteins with ChiSq pvalue =< 0.1. As a refresher, the ChiSq proportions test compares 23C and 29C protein total spectra/sample total spectra proportions).
- Node attribute file generated by R: all_sig0.1_pro_logFC_pval_abbrv_NodeAttb.csv
- Edge attribute file generate by R: edge_attb_semsim0.7_sig0.1.csv
- List of unique GO slim IDs from the analysis: sig0.1_GOslimIDs.csv
- I created a new network with edge_attb_semsim0.7_sig0.1.csv and all_sig0.1_pro_logFC_pval_abbrv_NodeAttb.csv in the same cytoscape session file as before OysterProteomicsNetworkAnalysis.cys
- I selected only GO slim terms and made a new network with these. I did tree layout, spread nodes out so their labels were not overlapping, and made this figure:

- The biggest clusters are metabolism, development, and transport, so I made subnetworks of these including their associated proteins. I colored nodes by their log foldchange and made these figures:

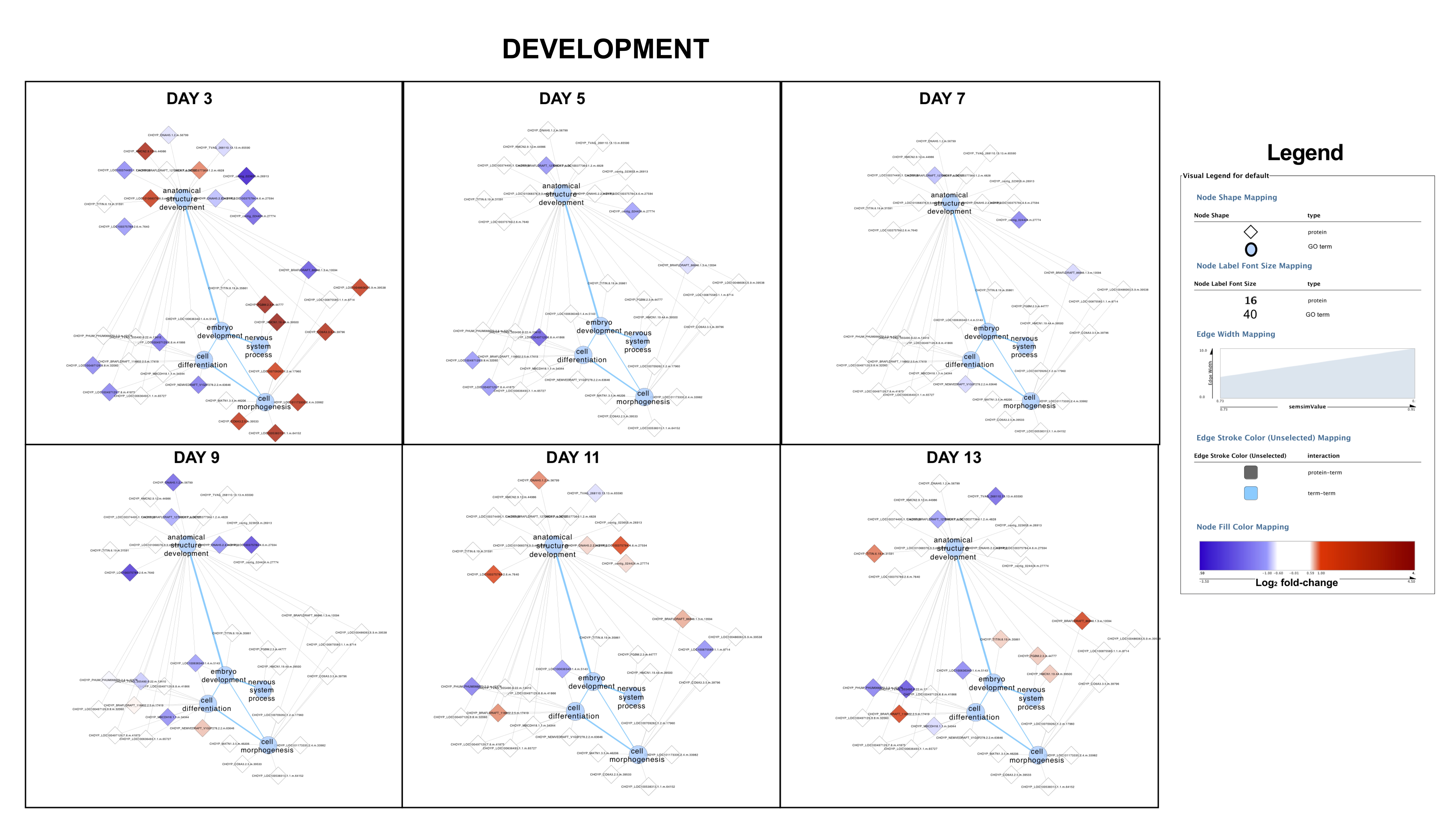
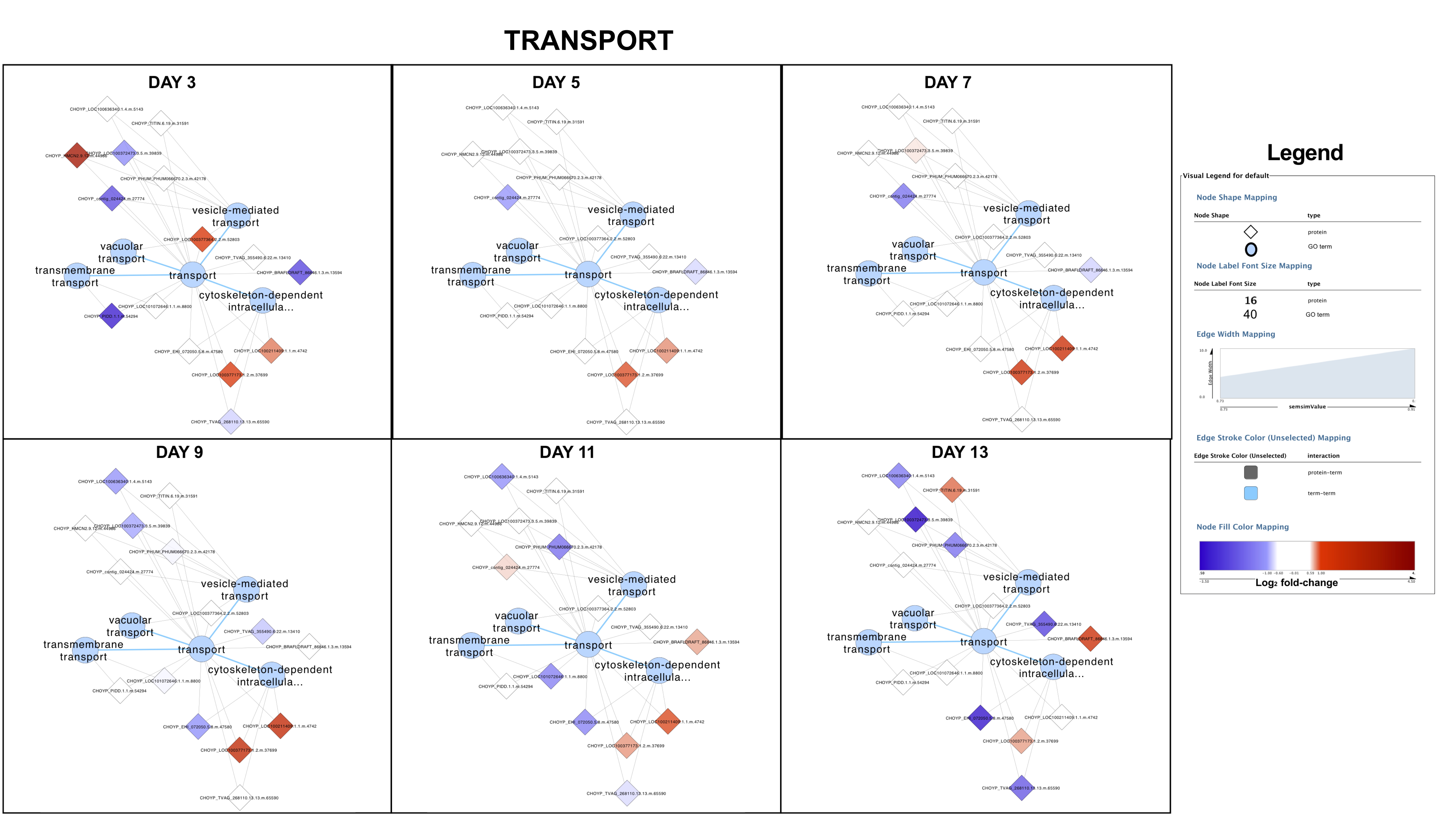
- these figures are all in this folder: https://github.com/shellywanamaker/OysterSeedProject/tree/master/analysis/UniprotAnnotations_NetworkAnalysis/SameDayFCtoTemp/ChiSqPval0.1proteins_GOsemsim_edges/ChiSqPval0.1proteins_GOsemsim_edges_Network_figures and you can zoom in on them there.
Conclusions:
- Temperature seems to affect the biological processes transport, metabolism, and development
- Regarding transport, a couple of proteins related to cytoskeleton-dependent intracellular transport are upregulated at 29C across almost all days while a few proteins related to vesicle mediated transport are downregulated especially on Day 13.
- Regarding metabolism, over time there is a general downregulation of proteins associated with different metabolic processes at 29C.
- Regarding development, early on at Day 3 there is an upregulation of proteins associated with cell morphogenesis and nervous system processes at 29C. Day 11 also shows more upregulated than downregulated development proteins at 29C
- revisiting past analysis, the PCA of technical replicates showed that 29C time points sometimes clustered with the succeeding 23C time points, suggesting the increased temperature could speed up development.
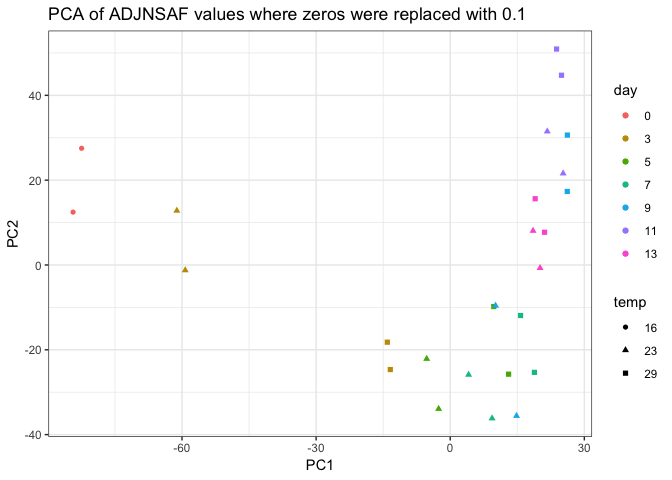
- This could be apparent in the ASCA PCA plot of the timextemperature interaction effect with most of the 23C time points being separated from most of the 29C time points along PC1. This could be more dramatic separation if I removed time point 0.

- revisiting phenotypic data Rhonda gathered, on Day 6, 29C seed was generally larger in size
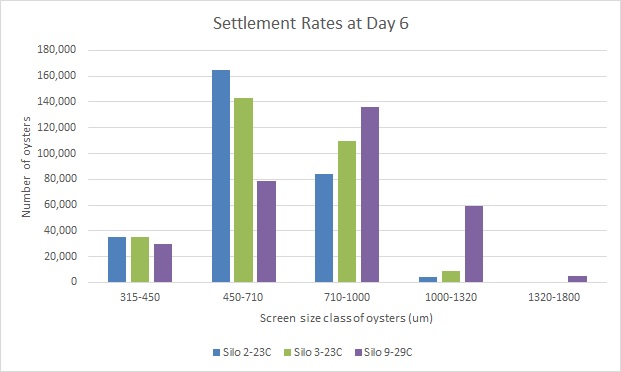 .
.
- revisiting past analysis, the PCA of technical replicates showed that 29C time points sometimes clustered with the succeeding 23C time points, suggesting the increased temperature could speed up development.
To do:
- Need to swap CHOYP names for gene names or protein names
- Dig into the lit about the proteins mentioned above so get a better idea of the implications of their temperature-altered abundance.
- Probably need to threshold the uniprot mapping by e-value since I haven’t done that yet
- Maybe try this mapping pipeline for molecular function GO terms
- see what people think about this network analysis
- It would be cool to visualize these networks as dynamic networks rather than each day side-by-side. And there seem to be a few Cytoscape apps for visualising dynamic networks