Diploid and triploid global DNA methylation
I re-did the global DNA methylation triploid/diploid pilot analysis in R to compare the results to the data Ronit and I generated Wednesday Feb 6.
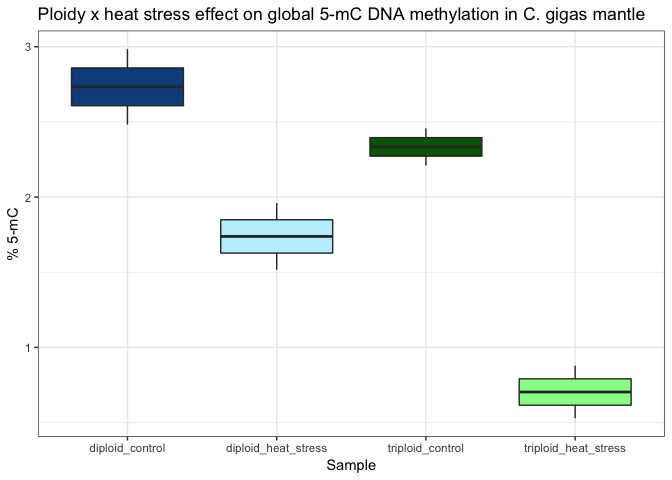
The pilot data showed diploid mantle has more methylation than triploid mantle. And there is a more dramatic decrease in global 5-mC methylation in response to heatstress (24hr at 27C, then 1hr at 40C) in triploid than in diploid mantle.
However, Ronit’s experiment with Ctenidia showed a different trend. Global methylation was not significantly different between diploids, triploids, or triploids exposed to heatstress (24hr at 27C). But diploids exposed to heatstress (24hr at 27C) showed a significant increase in methylation.
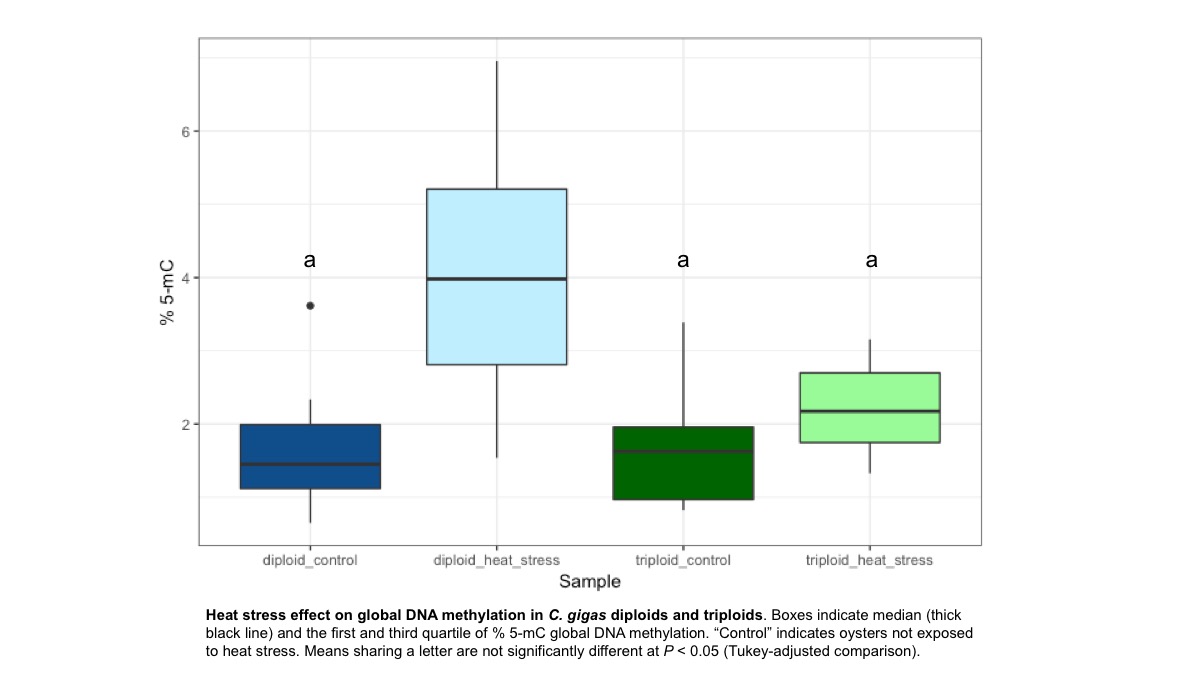
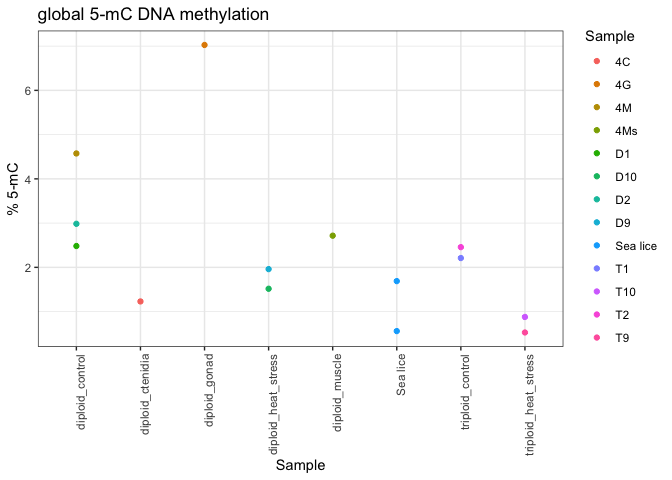
Since the pilot results are only from two biological replicates in each group, it’s not clear if these trends would persist if more mantle samples were processed.
The pilot data also showed a difference between tissue types, but again it’s not clear if these trends would still be seen if more samples were processed.
RNA from tanner crab hemolymph
Went through RNeasy kit with Grace, Steven, Laura, and Alana. Grace had 4 crab samples and Alana was preparing 4 juvenile oyster samples side-by-side.
After the gDNA eliminator column step, I took the gDNA eliminator column and eluted the gDNA off the column following the procedure below adapted from this researchgate post
 I ended up spinning at 4C since the room temp centrifuge was being used for the RNA extractions, and I ended up using 50-55C prewarmed AE since that’s was the heatblock was set at.
I ended up spinning at 4C since the room temp centrifuge was being used for the RNA extractions, and I ended up using 50-55C prewarmed AE since that’s was the heatblock was set at.
The DNA and RNA qubit results were as follows:

I saved the DNA samples in my DNA box in the -20C in rm 213.
We still aren’t sure whether the RNeasy columns are being overloaded or if the RNA in the samples is degraded. So next Grace is trying to isolate more RNA from sample 516 and will be testing 10uL vs. 50uL vs. 100uL of pellet slurry (starting material). This should hopefully determine the appropriate amount of starting material to use.