Amb-low versus super low-low
This analysis focuses on 8 samples:
- 4 x animals reared in ambient conditions for 23 days
- 4 x animals reared in super low pH conditions for 23 days
- following initial 23 day exposure, all animals were exposed to an ambient common garden indoors for 28 days, then an ambient common garden outdoors for 84 days, then underwent a secondary low pH exposure for 10 days after which they were preserved for methylome analysis.
- see Hollie’s experimental setup here
Compare coverage between v074 and v070
- Ran alignment to v074 using same settings as past alignments to v070
- mox scripts here: 20190806_BmrkPgenr074.sh
- output here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20190806_v074/
- Generate coverage reports for past analysis with v070
- Generate coverage reports for v074 because that part of the mox script 20190806_BmrkPgenr074.sh failed
- cov report mox script here: 20190807_BmrkCytCov074.sh
- ouput here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20190806_v074/
- Generate sorted bams for v074 analysis because that part of the mox script 20190806_BmrkPgenr074.sh failed
- sorted bams mox script here: 20190807_SortBams_Pgenr.sh
- output here: https://gannet.fish.washington.edu/metacarcinus/Pgenerosa/analyses/20190806_v074/
- Ran jupyter notebook to plot coverage for v074
- notebook here: 20190806_Pgnr_cmpr_cov.ipynb
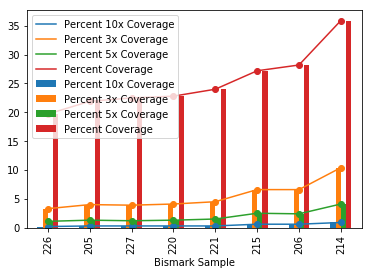
- didn’t get to run this on v070 yet so will do that later
- notebook here: 20190806_Pgnr_cmpr_cov.ipynb
Differential methylation analysis
- Updated old methylkit analysis on these samples with v070 alignments to output the DMR files
- R script here: Pgenr_JuviOARRBS_low_vs_ambient.Rmd
- R project here: methylkit_JuviPgenr_allData.Rproj
- DMR files here:
***found this article about cov.bases that Yaamini and Mac have previously mentioned. Doesn’t seem like it makes a difference if this is set to 0 or 1.
- Started running methylkit DMR analysis of v074 alignments on Emu
- copied bams over to Emu:
srlab@emu:~/GitHub/Shelly_Pgenerosa/analyses/JuviPgen_ALSL2lowd145/Get_DMRs_for_v074_alignments/dedup_bams$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20190806_v074/*_dedup.sorted.bam .
Rsync output from copying data from mox to gannet
[strigg@mox2 analyses]$ rsync --archive --progress --verbose /gscratch/scrubbed/strigg/analyses/20190806_v074 strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses
Password:
building file list ...
145 files to consider
20190806_v074/
20190806_v074/CHG_OB_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,204,054,204 100% 47.20MB/s 0:00:24 (xfr#1, to-chk=143/145)
20190806_v074/CHG_OB_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,828,394,312 100% 47.43MB/s 0:00:36 (xfr#2, to-chk=142/145)
20190806_v074/CHG_OB_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
2,527,215,131 100% 43.31MB/s 0:00:55 (xfr#3, to-chk=141/145)
20190806_v074/CHG_OB_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,776,103,415 100% 44.71MB/s 0:00:37 (xfr#4, to-chk=140/145)
20190806_v074/CHG_OB_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,284,122,063 100% 40.32MB/s 0:00:30 (xfr#5, to-chk=139/145)
20190806_v074/CHG_OB_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,361,954,058 100% 45.55MB/s 0:00:28 (xfr#6, to-chk=138/145)
20190806_v074/CHG_OB_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,064,328,288 100% 44.16MB/s 0:00:22 (xfr#7, to-chk=137/145)
20190806_v074/CHG_OB_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,215,726,502 100% 38.94MB/s 0:00:29 (xfr#8, to-chk=136/145)
20190806_v074/CHG_OT_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,192,628,629 100% 40.42MB/s 0:00:28 (xfr#9, to-chk=135/145)
20190806_v074/CHG_OT_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,807,003,959 100% 43.49MB/s 0:00:39 (xfr#10, to-chk=134/145)
20190806_v074/CHG_OT_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
2,509,107,895 100% 44.63MB/s 0:00:53 (xfr#11, to-chk=133/145)
20190806_v074/CHG_OT_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,754,028,073 100% 44.31MB/s 0:00:37 (xfr#12, to-chk=132/145)
20190806_v074/CHG_OT_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,264,092,387 100% 43.35MB/s 0:00:27 (xfr#13, to-chk=131/145)
20190806_v074/CHG_OT_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,349,340,180 100% 44.17MB/s 0:00:29 (xfr#14, to-chk=130/145)
20190806_v074/CHG_OT_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,047,667,263 100% 45.74MB/s 0:00:21 (xfr#15, to-chk=129/145)
20190806_v074/CHG_OT_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,200,576,000 100% 43.86MB/s 0:00:26 (xfr#16, to-chk=128/145)
20190806_v074/CHH_OB_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,658,775,548 100% 43.28MB/s 0:01:20 (xfr#17, to-chk=127/145)
20190806_v074/CHH_OB_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
5,518,727,115 100% 44.28MB/s 0:01:58 (xfr#18, to-chk=126/145)
20190806_v074/CHH_OB_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
7,848,248,612 100% 43.94MB/s 0:02:50 (xfr#19, to-chk=125/145)
20190806_v074/CHH_OB_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
5,472,810,448 100% 41.03MB/s 0:02:07 (xfr#20, to-chk=124/145)
20190806_v074/CHH_OB_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
4,100,955,167 100% 45.49MB/s 0:01:25 (xfr#21, to-chk=123/145)
20190806_v074/CHH_OB_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
4,314,283,667 100% 43.87MB/s 0:01:33 (xfr#22, to-chk=122/145)
20190806_v074/CHH_OB_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,392,446,875 100% 45.73MB/s 0:01:10 (xfr#23, to-chk=121/145)
20190806_v074/CHH_OB_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,897,193,762 100% 43.08MB/s 0:01:26 (xfr#24, to-chk=120/145)
20190806_v074/CHH_OT_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,628,543,634 100% 45.75MB/s 0:01:15 (xfr#25, to-chk=119/145)
20190806_v074/CHH_OT_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
5,473,629,239 100% 42.08MB/s 0:02:04 (xfr#26, to-chk=118/145)
20190806_v074/CHH_OT_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
7,798,653,297 100% 44.89MB/s 0:02:45 (xfr#27, to-chk=117/145)
20190806_v074/CHH_OT_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
5,421,814,698 100% 42.69MB/s 0:02:01 (xfr#28, to-chk=116/145)
20190806_v074/CHH_OT_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
4,061,692,626 100% 43.76MB/s 0:01:28 (xfr#29, to-chk=115/145)
20190806_v074/CHH_OT_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
4,281,496,520 100% 42.04MB/s 0:01:37 (xfr#30, to-chk=114/145)
20190806_v074/CHH_OT_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,358,482,102 100% 46.16MB/s 0:01:09 (xfr#31, to-chk=113/145)
20190806_v074/CHH_OT_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
3,871,442,663 100% 44.25MB/s 0:01:23 (xfr#32, to-chk=112/145)
20190806_v074/CpG_OB_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
794,708,749 100% 43.74MB/s 0:00:17 (xfr#33, to-chk=111/145)
20190806_v074/CpG_OB_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,184,493,224 100% 44.26MB/s 0:00:25 (xfr#34, to-chk=110/145)
20190806_v074/CpG_OB_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,658,009,553 100% 44.12MB/s 0:00:35 (xfr#35, to-chk=109/145)
20190806_v074/CpG_OB_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,141,504,240 100% 39.77MB/s 0:00:27 (xfr#36, to-chk=108/145)
20190806_v074/CpG_OB_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
823,494,640 100% 41.57MB/s 0:00:18 (xfr#37, to-chk=107/145)
20190806_v074/CpG_OB_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
874,287,987 100% 35.48MB/s 0:00:23 (xfr#38, to-chk=106/145)
20190806_v074/CpG_OB_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
681,089,281 100% 44.12MB/s 0:00:14 (xfr#39, to-chk=105/145)
20190806_v074/CpG_OB_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
785,170,609 100% 42.86MB/s 0:00:17 (xfr#40, to-chk=104/145)
20190806_v074/CpG_OT_EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
782,298,004 100% 44.11MB/s 0:00:16 (xfr#41, to-chk=103/145)
20190806_v074/CpG_OT_EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,164,180,284 100% 44.09MB/s 0:00:25 (xfr#42, to-chk=102/145)
20190806_v074/CpG_OT_EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,635,665,583 100% 45.41MB/s 0:00:34 (xfr#43, to-chk=101/145)
20190806_v074/CpG_OT_EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
1,122,011,753 100% 40.03MB/s 0:00:26 (xfr#44, to-chk=100/145)
20190806_v074/CpG_OT_EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
806,463,320 100% 40.98MB/s 0:00:18 (xfr#45, to-chk=99/145)
20190806_v074/CpG_OT_EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
861,523,166 100% 44.34MB/s 0:00:18 (xfr#46, to-chk=98/145)
20190806_v074/CpG_OT_EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
667,210,945 100% 39.07MB/s 0:00:16 (xfr#47, to-chk=97/145)
20190806_v074/CpG_OT_EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.txt
769,840,235 100% 44.94MB/s 0:00:16 (xfr#48, to-chk=96/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,790 100% 913.60kB/s 0:00:00 (xfr#49, to-chk=95/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,010 100% 4.91kB/s 0:00:00 (xfr#50, to-chk=94/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.bam
1,415,720,462 100% 45.33MB/s 0:00:29 (xfr#51, to-chk=93/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,688 100% 20.42kB/s 0:00:00 (xfr#52, to-chk=92/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,043,385,336 100% 44.58MB/s 0:00:22 (xfr#53, to-chk=91/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
43,234,484 100% 32.80MB/s 0:00:01 (xfr#54, to-chk=90/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
37,557,825 100% 33.44MB/s 0:00:01 (xfr#55, to-chk=89/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
879 100% 11.60kB/s 0:00:00 (xfr#56, to-chk=88/145)
20190806_v074/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
351 100% 3.33kB/s 0:00:00 (xfr#57, to-chk=87/145)
20190806_v074/EPI-205_S26_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
131,013,238 100% 42.58MB/s 0:00:02 (xfr#58, to-chk=86/145)
20190806_v074/EPI-205_S26_L004_dedup.sorted.bam
875,133,911 100% 39.16MB/s 0:00:21 (xfr#59, to-chk=85/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,855 100% 773.33kB/s 0:00:00 (xfr#60, to-chk=84/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,012 100% 4.14kB/s 0:00:00 (xfr#61, to-chk=83/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.bam
2,032,293,966 100% 45.40MB/s 0:00:42 (xfr#62, to-chk=82/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,941 100% 22.30kB/s 0:00:00 (xfr#63, to-chk=81/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,537,982,344 100% 42.28MB/s 0:00:34 (xfr#64, to-chk=80/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
55,459,884 100% 28.45MB/s 0:00:01 (xfr#65, to-chk=79/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
49,266,400 100% 24.98MB/s 0:00:01 (xfr#66, to-chk=78/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
879 100% 0.98kB/s 0:00:00 (xfr#67, to-chk=77/145)
20190806_v074/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
351 100% 0.39kB/s 0:00:00 (xfr#68, to-chk=76/145)
20190806_v074/EPI-206_S27_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
133,539,018 100% 34.38MB/s 0:00:03 (xfr#69, to-chk=75/145)
20190806_v074/EPI-206_S27_L004_dedup.sorted.bam
1,252,773,646 100% 44.58MB/s 0:00:26 (xfr#70, to-chk=74/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,982 100% 416.56kB/s 0:00:00 (xfr#71, to-chk=73/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,013 100% 2.23kB/s 0:00:00 (xfr#72, to-chk=72/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.bam
2,921,053,627 100% 45.53MB/s 0:01:01 (xfr#73, to-chk=71/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
17,235 100% 180.98kB/s 0:00:00 (xfr#74, to-chk=70/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
2,135,882,869 100% 45.79MB/s 0:00:44 (xfr#75, to-chk=69/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
70,347,871 100% 30.13MB/s 0:00:02 (xfr#76, to-chk=68/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
63,718,431 100% 37.28MB/s 0:00:01 (xfr#77, to-chk=67/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
882 100% 1.36kB/s 0:00:00 (xfr#78, to-chk=66/145)
20190806_v074/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
353 100% 0.54kB/s 0:00:00 (xfr#79, to-chk=65/145)
20190806_v074/EPI-214_S30_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
136,509,692 100% 35.45MB/s 0:00:03 (xfr#80, to-chk=64/145)
20190806_v074/EPI-214_S30_L004_dedup.sorted.bam
1,701,814,404 100% 41.84MB/s 0:00:38 (xfr#81, to-chk=63/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,857 100% 973.52kB/s 0:00:00 (xfr#82, to-chk=62/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,013 100% 5.17kB/s 0:00:00 (xfr#83, to-chk=61/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.bam
2,126,356,975 100% 42.82MB/s 0:00:47 (xfr#84, to-chk=60/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,939 100% 120.74kB/s 0:00:00 (xfr#85, to-chk=59/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,503,368,044 100% 45.64MB/s 0:00:31 (xfr#86, to-chk=58/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
53,761,313 100% 34.14MB/s 0:00:01 (xfr#87, to-chk=57/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
47,824,286 100% 30.12MB/s 0:00:01 (xfr#88, to-chk=56/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
879 100% 1.67kB/s 0:00:00 (xfr#89, to-chk=55/145)
20190806_v074/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
352 100% 0.65kB/s 0:00:00 (xfr#90, to-chk=54/145)
20190806_v074/EPI-215_S31_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
133,434,500 100% 38.08MB/s 0:00:03 (xfr#91, to-chk=53/145)
20190806_v074/EPI-215_S31_L004_dedup.sorted.bam
1,217,722,626 100% 43.99MB/s 0:00:26 (xfr#92, to-chk=52/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,799 100% 840.53kB/s 0:00:00 (xfr#93, to-chk=51/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,011 100% 4.46kB/s 0:00:00 (xfr#94, to-chk=50/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.bam
1,646,052,864 100% 44.74MB/s 0:00:35 (xfr#95, to-chk=49/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,786 100% 145.07kB/s 0:00:00 (xfr#96, to-chk=48/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,099,806,886 100% 45.55MB/s 0:00:23 (xfr#97, to-chk=47/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
44,928,221 100% 42.85MB/s 0:00:01 (xfr#98, to-chk=46/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
39,035,250 100% 37.95MB/s 0:00:00 (xfr#99, to-chk=45/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
879 100% 0.87kB/s 0:00:00 (xfr#100, to-chk=44/145)
20190806_v074/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
351 100% 0.35kB/s 0:00:00 (xfr#101, to-chk=43/145)
20190806_v074/EPI-220_S32_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
131,227,349 100% 33.24MB/s 0:00:03 (xfr#102, to-chk=42/145)
20190806_v074/EPI-220_S32_L004_dedup.sorted.bam
916,828,880 100% 36.49MB/s 0:00:23 (xfr#103, to-chk=41/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,808 100% 675.30kB/s 0:00:00 (xfr#104, to-chk=40/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,011 100% 3.64kB/s 0:00:00 (xfr#105, to-chk=39/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.bam
1,637,659,455 100% 45.28MB/s 0:00:34 (xfr#106, to-chk=38/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,797 100% 32.48kB/s 0:00:00 (xfr#107, to-chk=37/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,164,916,000 100% 40.76MB/s 0:00:27 (xfr#108, to-chk=36/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
47,256,049 100% 21.85MB/s 0:00:02 (xfr#109, to-chk=35/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
41,216,575 100% 33.86MB/s 0:00:01 (xfr#110, to-chk=34/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
879 100% 5.33kB/s 0:00:00 (xfr#111, to-chk=33/145)
20190806_v074/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
351 100% 1.85kB/s 0:00:00 (xfr#112, to-chk=32/145)
20190806_v074/EPI-221_S33_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
131,667,804 100% 35.43MB/s 0:00:03 (xfr#113, to-chk=31/145)
20190806_v074/EPI-221_S33_L004_dedup.sorted.bam
971,611,127 100% 39.40MB/s 0:00:23 (xfr#114, to-chk=30/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,720 100% 670.07kB/s 0:00:00 (xfr#115, to-chk=29/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,010 100% 3.61kB/s 0:00:00 (xfr#116, to-chk=28/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.bam
1,252,453,489 100% 40.06MB/s 0:00:29 (xfr#117, to-chk=27/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,666 100% 20.02kB/s 0:00:00 (xfr#118, to-chk=26/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
903,432,465 100% 40.24MB/s 0:00:21 (xfr#119, to-chk=25/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
38,858,828 100% 26.25MB/s 0:00:01 (xfr#120, to-chk=24/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
33,470,064 100% 28.27MB/s 0:00:01 (xfr#121, to-chk=23/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
876 100% 6.74kB/s 0:00:00 (xfr#122, to-chk=22/145)
20190806_v074/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
350 100% 2.67kB/s 0:00:00 (xfr#123, to-chk=21/145)
20190806_v074/EPI-226_S34_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
130,079,212 100% 31.25MB/s 0:00:03 (xfr#124, to-chk=20/145)
20190806_v074/EPI-226_S34_L004_dedup.sorted.bam
753,681,291 100% 36.42MB/s 0:00:19 (xfr#125, to-chk=19/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_PE_report.html
365,777 100% 404.53kB/s 0:00:00 (xfr#126, to-chk=18/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_PE_report.txt
2,009 100% 2.15kB/s 0:00:00 (xfr#127, to-chk=17/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.bam
1,441,673,440 100% 43.08MB/s 0:00:31 (xfr#128, to-chk=16/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.M-bias.txt
16,749 100% 18.50kB/s 0:00:00 (xfr#129, to-chk=15/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bam
1,039,075,931 100% 44.06MB/s 0:00:22 (xfr#130, to-chk=14/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bedGraph.gz
44,195,627 100% 30.02MB/s 0:00:01 (xfr#131, to-chk=13/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz
38,291,186 100% 30.06MB/s 0:00:01 (xfr#132, to-chk=12/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated_splitting_report.txt
878 100% 3.95kB/s 0:00:00 (xfr#133, to-chk=11/145)
20190806_v074/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplication_report.txt
351 100% 1.54kB/s 0:00:00 (xfr#134, to-chk=10/145)
20190806_v074/EPI-227_S35_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
131,022,369 100% 32.29MB/s 0:00:03 (xfr#135, to-chk=9/145)
20190806_v074/EPI-227_S35_L004_dedup.sorted.bam
871,297,403 100% 39.97MB/s 0:00:20 (xfr#136, to-chk=8/145)
20190806_v074/bismark_summary_report.html
272,185 100% 373.85kB/s 0:00:00 (xfr#137, to-chk=7/145)
20190806_v074/bismark_summary_report.txt
1,545 100% 2.12kB/s 0:00:00 (xfr#138, to-chk=6/145)
20190806_v074/bme.err
208,573 100% 279.02kB/s 0:00:00 (xfr#139, to-chk=5/145)
20190806_v074/dedup.err
5,869 100% 7.67kB/s 0:00:00 (xfr#140, to-chk=4/145)
20190806_v074/mapping_dedup_summary.txt
704 100% 0.91kB/s 0:00:00 (xfr#141, to-chk=3/145)
20190806_v074/slurm-1154897.out
189,204 100% 233.00kB/s 0:00:00 (xfr#142, to-chk=2/145)
20190806_v074/slurm-1156605.out
505 100% 0.62kB/s 0:00:00 (xfr#143, to-chk=1/145)
20190806_v074/slurm-1156661.out
39,001 100% 46.91kB/s 0:00:00 (xfr#144, to-chk=0/145)
sent 151,528,699,160 bytes received 3,198 bytes 46,036,367.11 bytes/sec
total size is 151,510,187,727 speedup is 1.00
[strigg@mox2 analyses]$ rsync --archive --progress --verbose /gscratch/srlab/strigg/jobs/ strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/mox_jobs/
Password:
building file list ...
32 files to consider
./
20190806_BmrkPgenr074.sh
3,445 100% 2.62MB/s 0:00:00 (xfr#1, to-chk=4/32)
20190807_BismarkCoverage2Cytosine.sh
1,096 100% 1.05MB/s 0:00:00 (xfr#2, to-chk=2/32)
20190807_BmrkCytCov074.sh
1,042 100% 1017.58kB/s 0:00:00 (xfr#3, to-chk=1/32)
20190807_SortBams_Pgenr.sh
906 100% 884.77kB/s 0:00:00 (xfr#4, to-chk=0/32)
sent 6,053 bytes received 310 bytes 1,818.00 bytes/sec
total size is 116,252 speedup is 18.27
[strigg@mox2 20190807_Nov1_Pgenr_CytCov]$ rsync --archive --progress --verbose --exclude "data" /gscratch/scrubbed/strigg/analyses/20190807_Nov1_Pgenr_CytCov strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses
Password:
building file list ...
10 files to consider
20190807_Nov1_Pgenr_CytCov/
20190807_Nov1_Pgenr_CytCov/EPI-205_S26_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
327,334,629 100% 46.42MB/s 0:00:06 (xfr#1, to-chk=8/10)
20190807_Nov1_Pgenr_CytCov/EPI-206_S27_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
330,554,417 100% 41.89MB/s 0:00:07 (xfr#2, to-chk=7/10)
20190807_Nov1_Pgenr_CytCov/EPI-214_S30_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
334,319,152 100% 41.36MB/s 0:00:07 (xfr#3, to-chk=6/10)
20190807_Nov1_Pgenr_CytCov/EPI-215_S31_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
330,368,526 100% 41.65MB/s 0:00:07 (xfr#4, to-chk=5/10)
20190807_Nov1_Pgenr_CytCov/EPI-220_S32_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
327,572,369 100% 42.38MB/s 0:00:07 (xfr#5, to-chk=4/10)
20190807_Nov1_Pgenr_CytCov/EPI-221_S33_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
328,162,365 100% 43.61MB/s 0:00:07 (xfr#6, to-chk=3/10)
20190807_Nov1_Pgenr_CytCov/EPI-226_S34_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
326,096,618 100% 34.42MB/s 0:00:09 (xfr#7, to-chk=2/10)
20190807_Nov1_Pgenr_CytCov/EPI-227_S35_L004_cytosine_CpG_cov_report.CpG_report.txt.gz
327,322,006 100% 40.39MB/s 0:00:07 (xfr#8, to-chk=1/10)
20190807_Nov1_Pgenr_CytCov/slurm-1156627.out
400,017,493 100% 40.20MB/s 0:00:09 (xfr#9, to-chk=0/10)
sent 3,032,118,715 bytes received 228 bytes 44,920,280.64 bytes/sec
total size is 3,031,747,575 speedup is 1.00
ouput from copying data from gannet to mox
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-205_S26_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 45MB 40.9MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-206_S27_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 59MB 42.8MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-214_S30_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 77MB 39.2MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-215_S31_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 57MB 41.0MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-220_S32_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 47MB 40.6MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-221_S33_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 49MB 37.0MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-226_S34_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 40MB 39.3MB/s 00:01
[strigg@mox2 data]$ scp strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Pgenerosa/analyses/20181101/EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz .
Password:
EPI-227_S35_L004_R1_001_val_1_bismark_bt2_pe.deduplicated.bismark.cov.gz 100% 46MB 41.3MB/s 00:01