Vitellogenin detection in geoduck and RRBS analysis of juvenile geoduck exposed to low pH
-
updated author list and re-approved Sci Reports manuscript
-
Corrected Pgenr Mox script to run Bismark on subset of Holly’s BS data, and submitted job (in queue because nodes are being used)
-
Submitted Bismark job on full Cvirg dataset on Mox with analysis time extended to 10 days because the previous time (1d15h) was not enough to finish the job, hence why it timed out.
-
copied over incomplete analysis to Cvirg metacarcinus folder on Gannet to try methylkit analysis while waiting for job to complete: pwd /Volumes/web/metacarcinus/Cvirginica
rsync –archive –progress –verbose strigg@mox.hyak.uw.edu:/gscratch/srlab/strigg/analyses/20181004 .
-
-
lab meeting @1pm
-
ELISA detection of vitellogenin in Geoduck:
- emailed sgarcia@uabc.edu.mx on 10-12-2018 about C. corteziensis Vtg/Vn polyclonal antibody from Fabiola’s paper
- Fabiola’s paper: compared translated mRNA sequences of Vtg/Vn from Panopea globosa and Crassostrea corteziensis to see if C. corteziensis VTG/Vn antibody would be compatible. I wanted to see the percent mapping between these protein sequences to get a sense of how much alignment and what domain homology is needed, but I couldn’t find these sequences in the Genbank as the paper says.
- Contacted TECO about their multispecies ELISA kit for vitellogenin detection in fish:
- Elizabeth Mihal @ TECO said (Oct 11, 2018): “Unfortunately, we do not have any indication that the Multispecies VTG kit would cross react with shellfish. I have attached a cross reactivity chart for your reference where you can see what species of fish have been shown to cross react with each kit. It is my understanding that the VTG in shellfish is too divergent from the VTG of Japanese rice fish, Australian rainbowfish, cod and flounder so it would make for an inappropriate sample type.” Some info testing the kit compatibility with various fish species has been published
- tried general protein alignment of Geoduck VTG with BLASTP
- made fasta file with Geoduck vitellogenin sequence from Emma’s paper (cds.comp144315_c0_seqI_m50928)
- performed BLASTP analysis using BLAST website
- alignment graphical summary
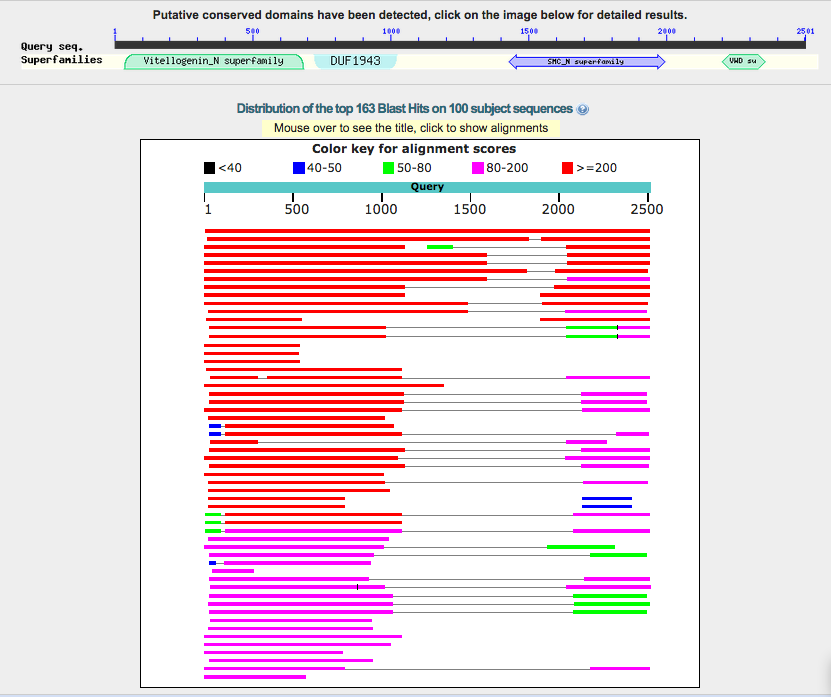
- bowser view of alignments
- top alignments
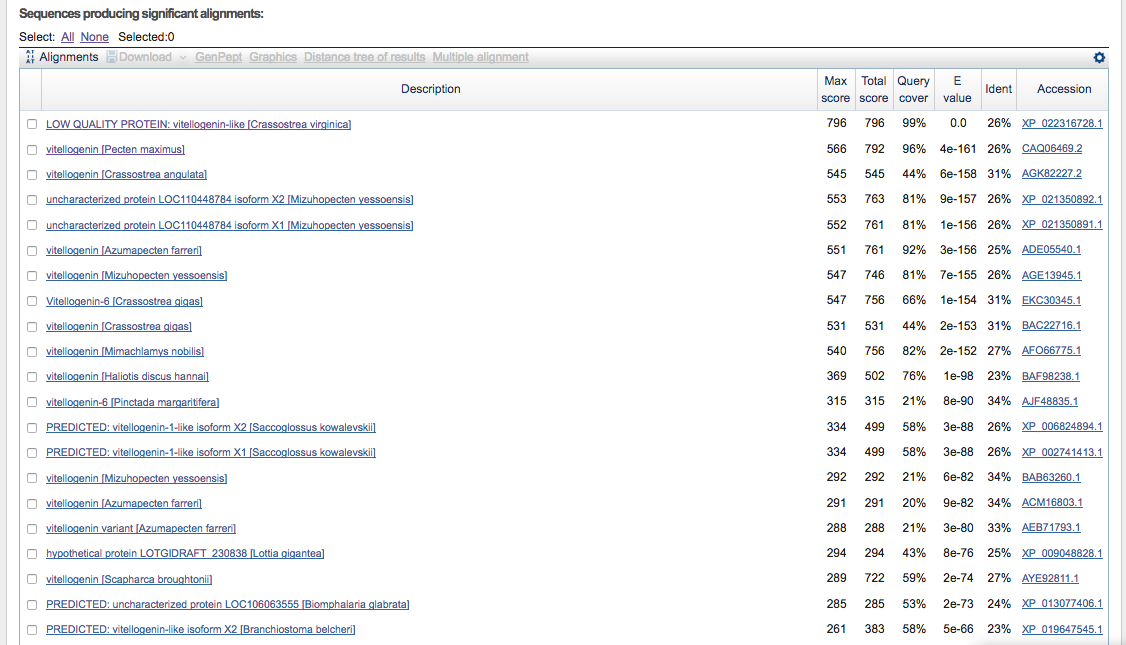
- top hit
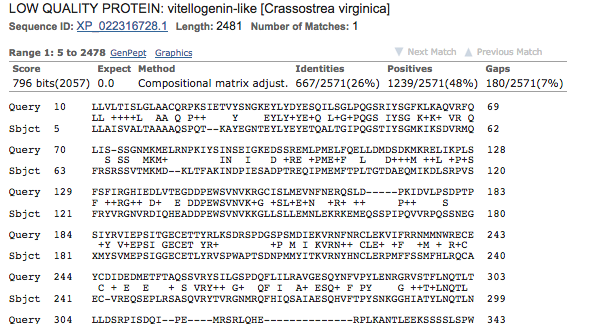
- Organism summary
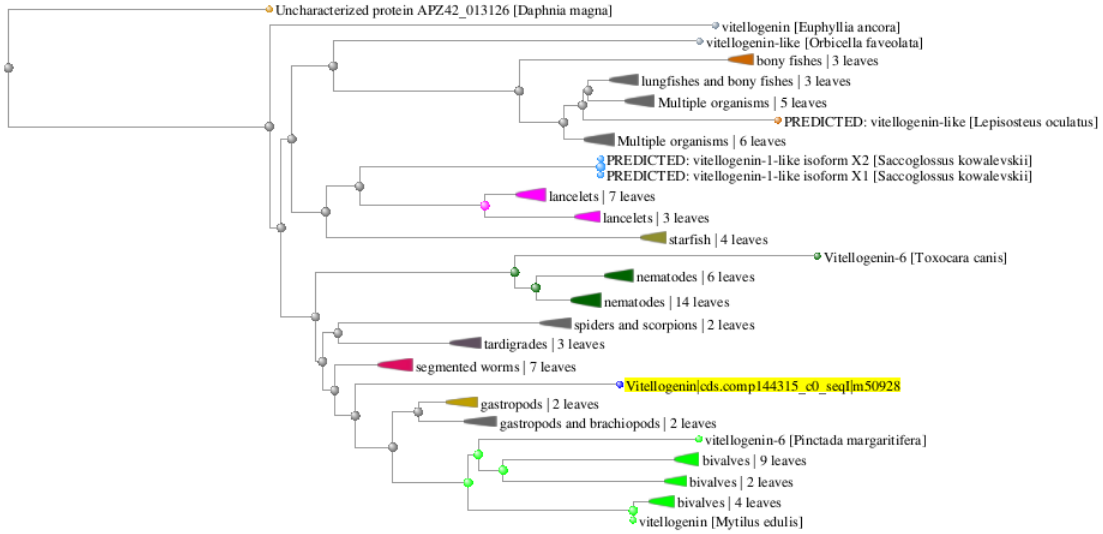
- Phylogenic tree
- alignment graphical summary
Conclusions: Pgenr VTG aligns best with VTG from other bivalves; BLASTp returned no species listed in TECO kits
- tried protein alignment of Geoduck VTG to VTG from species used in TECO kit:
- first tried aligning first Geoduck VTG domain which BLAST identified as ‘Vitellogenin N superfamily’ to species specific data by entering organism in search set options, figuring this domain was most conserved.
- alignments to zebrafish (which TECO kit works on)
- alignments to Japanese rice fish
- I compared these alignments to the alignment of two fish species (zebrafish and goldfish) that show medium crossreactivity with the ELISA kit
- next made fasta file with different species with detectable VTG in TECO kit
- used BLASTp with option to align multiple sequences and uploaded the Pgenr fasta file and the TECO species fasta file
- Phylogenic tree
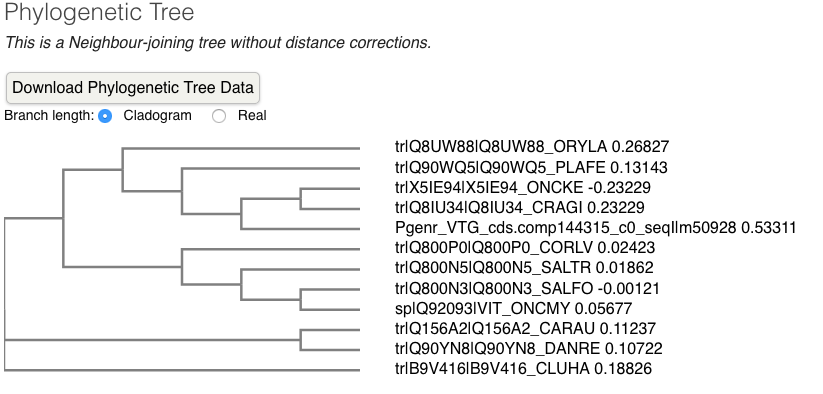
- alignments
- Phylogenic tree
- used clustal omega to perform multiple alignment with default settings
- first tried aligning first Geoduck VTG domain which BLAST identified as ‘Vitellogenin N superfamily’ to species specific data by entering organism in search set options, figuring this domain was most conserved.
Conclusions: TECO species VTGs align much better to each other than Pgenr VTG aligns with TECO species VTG. Pgenr VTG is more similar to C. gigas than to any TECO fish species and Pgenr VTG is most likely too divergent from TECO fish species.
mRNA expression may be the better way to go
- emailed sgarcia@uabc.edu.mx on 10-12-2018 about C. corteziensis Vtg/Vn polyclonal antibody from Fabiola’s paper