Received salmon sea lice methylome data!!
copy data to Gannet metacarcinus folder
- I received this url from the sequencing core
- I mounted gannet via finder -> connect to server
- I installed Globus Connect Personal on Ostrich and gave the Globus app writing permissions to my metacarcinus folder on Gannet
- I navigated to my Globus account file manager section via the provided url and could see my sequencing data “UW_Trigg_190718.tar.gz” 148.22GB.
- I selected the “Transfer of Sync to…” option, entered the computer name “Ostrich” which I entered when setting up Globus Connect Personal app, entered the path “/Volumes/web/metacarcinus/Salmo_Calig/FASTQS/”. Then clicked start.
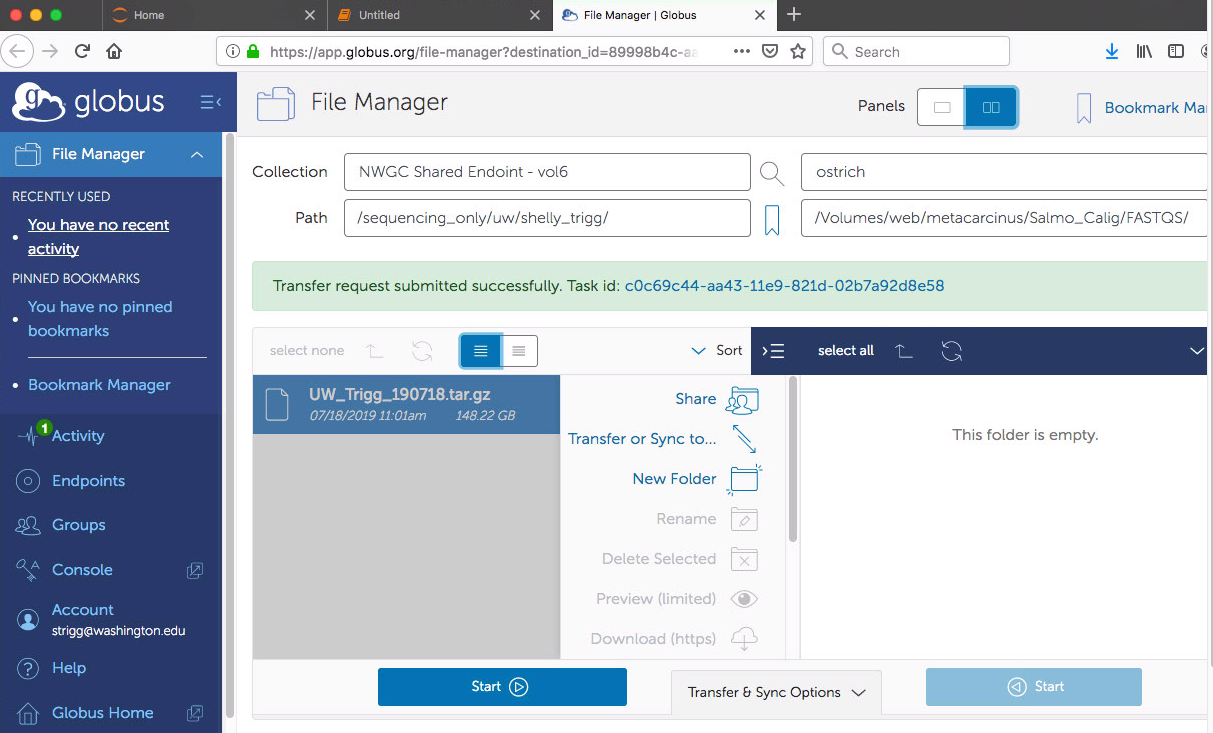
- I navigated to the Activity section of Globus and could see the info being transferred (26GB in 10min)
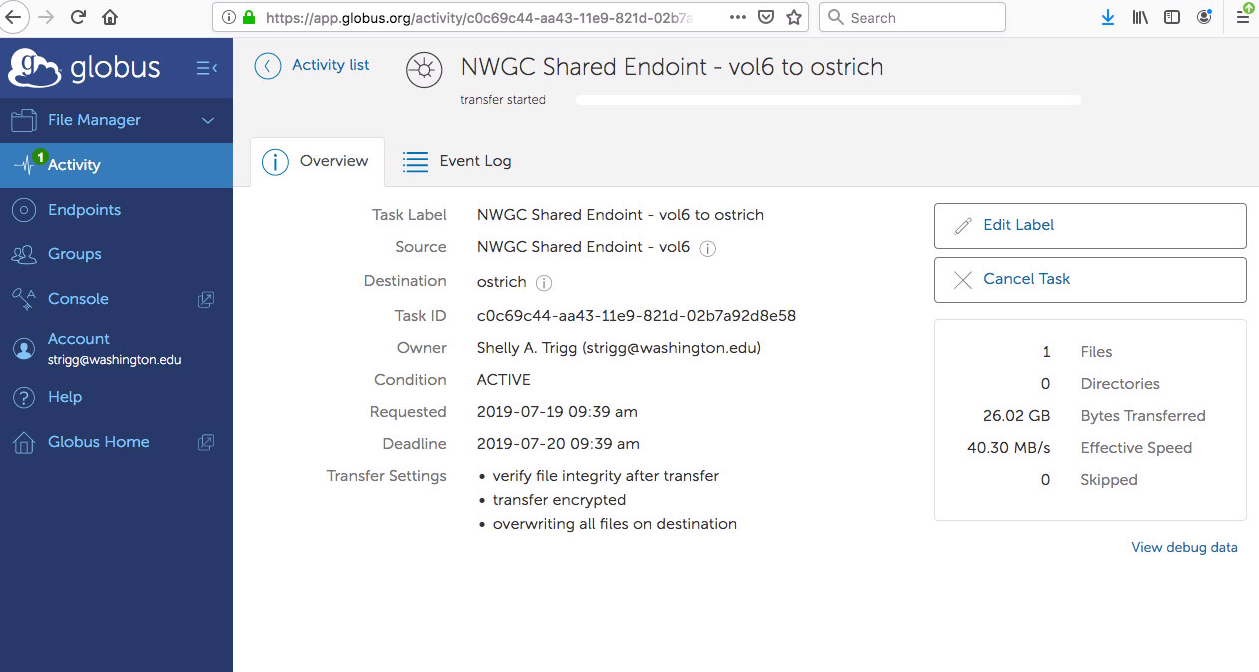
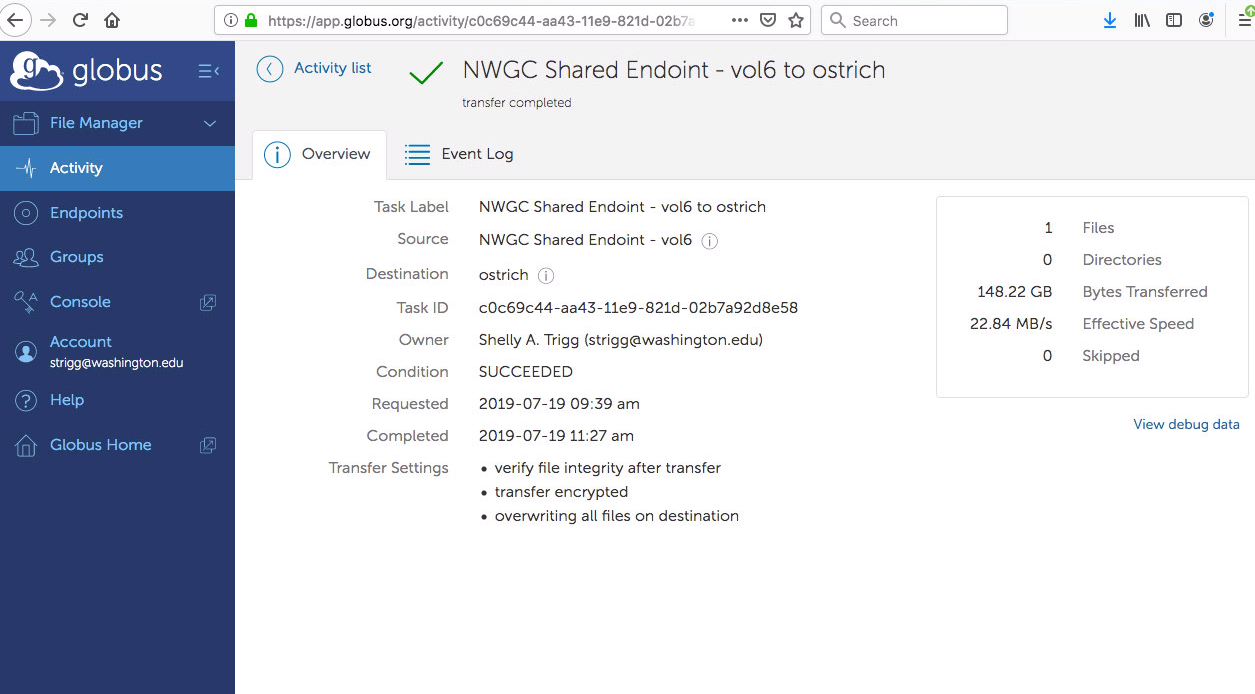
- after a couple hours, I received an email notification and could see in the status section that the transfer had completed successfully
copy data from Gannet to mox
ran this command:
rsync --archive --progress --verbose strigg@ostrich.fish.washington.edu:/Volumes/web/metacarcinus/Salmo_Calig/FASTQS/UW_Trigg_190718.tar.gz /gscratch/srlab/strigg/data/Salmo_Calig/FASTQS/
I decided to make two species folders rather than have them combined, and wanted to remove the data that was not Salmon or sea lice fastqs, so ran the following commands:
tar -xvf UW_Trigg_190718.tar.gz
cd UW_Trigg_190718_done/
rm -r Reports/
rm -r Stats/
rm -r Undetermined_S0_L00*
cd ../../../
mkdir Caligus
mv Salmo_Calig/ Ssalar
cd Ssalar/
cd FASTQS/
cd ..
cd Caligus/
mkdir FASTQS
mv ../Ssalar/FASTQS/UW_Trigg_190718_done/Sealice_F* .
mv *.gz FASTQS/
rm UW_Trigg_190718.tar.gz
copy data from Gannet to owl
- need to ask sam how to do this
- need to make new species directories for this data
- C_rogercresseyi
- S_salar
- made this meta data to add to the Nightingales database google sheet
run fastqc and trim
- prepared scripts (Salmon: /gscratch/srlab/strigg/jobs/20190719_FASTQC_ADPTRIM_Ssalar.sh and lice: /gscratch/srlab/strigg/jobs/20190719_FASTQC_ADPTRIM_Caligus.sh) to do this on Mox but didn’t work because multiqc and cutadapt programs could not be found as I had them. See github issue #712